Microdiversity of deep-sea Bacillales isolated from Tyrrhenian sea sediments as revealed by ARISA, 16S rRNA gene sequencing and BOX-PCR fingerprinting
- PMID: 24005887
- PMCID: PMC4070960
- DOI: 10.1264/jsme2.me13013
Microdiversity of deep-sea Bacillales isolated from Tyrrhenian sea sediments as revealed by ARISA, 16S rRNA gene sequencing and BOX-PCR fingerprinting
Abstract
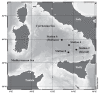
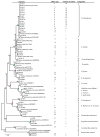
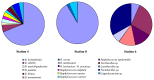
With respect to their terrestrial relatives, marine Bacillales have not been sufficiently investigated. In this report, the diversity of deep-sea Bacillales, isolated from seamount and non-seamount stations at 3,425 to 3,580 m depth in the Tyrrhenian Sea, was investigated using PCR fingerprinting and 16S rRNA sequence analysis. The isolate collection (n=120) was de-replicated by automated ribosomal intergenic spacer analysis (ARISA), and phylogenetic diversity was analyzed by 16S rRNA gene sequencing of representatives of each ARISA haplotype (n=37). Phylogenetic analysis of isolates showed their affiliation to six different genera of low G+C% content Gram-positive Bacillales: Bacillus, Staphylococcus, Exiguobacterium, Paenibacillus, Lysinibacillus and Terribacillus. Bacillus was the dominant genus represented by the species B. licheniformis, B. pumilus, B. subtilis, B. amyloliquefaciens and B. firmus, typically isolated from marine sediments. The most abundant species in the collection was B. licheniformis (n=85), which showed seven distinct ARISA haplotypes with haplotype H8 being the most dominant since it was identified by 63 isolates. The application of BOX-PCR fingerprinting to the B. licheniformis sub-collection allowed their separation into five distinct BOX genotypes, suggesting a high level of intraspecies diversity among marine B. licheniformis strains. This species also exhibited distinct strain distribution between seamount and non-seamount stations and was shown to be highly prevalent in non-seamount stations. This study revealed the great microdiversity of marine Bacillales and contributes to understanding the biogeographic distribution of marine bacteria in deep-sea sediments.
Figures
Similar articles
-
Diversity, ecological distribution and biotechnological potential of Actinobacteria inhabiting seamounts and non-seamounts in the Tyrrhenian Sea.Microbiol Res. 2016 May-Jun;186-187:71-80. doi: 10.1016/j.micres.2016.03.006. Epub 2016 Apr 1. Microbiol Res. 2016. PMID: 27242145
-
Diversity and phylogeny of culturable spore-forming Bacilli isolated from marine sediments.J Basic Microbiol. 2009 Sep;49 Suppl 1:S13-23. doi: 10.1002/jobm.200800306. J Basic Microbiol. 2009. PMID: 19322832
-
Gammaproteobacteria occurrence and microdiversity in Tyrrhenian Sea sediments as revealed by cultivation-dependent and -independent approaches.Syst Appl Microbiol. 2010 Jun;33(4):222-31. doi: 10.1016/j.syapm.2010.02.005. Epub 2010 Apr 22. Syst Appl Microbiol. 2010. PMID: 20413241
-
Genotypic characterization of Brochothrix spp. isolated from meat, poultry and fish.Lett Appl Microbiol. 2010 Sep;51(3):245-51. doi: 10.1111/j.1472-765X.2010.02886.x. Lett Appl Microbiol. 2010. PMID: 20716107
-
Bacillales: From Taxonomy to Biotechnological and Industrial Perspectives.Microorganisms. 2022 Nov 28;10(12):2355. doi: 10.3390/microorganisms10122355. Microorganisms. 2022. PMID: 36557608 Free PMC article. Review.
Cited by
-
Fungal diversity in deep-sea sediments from the Magellan seamounts as revealed by a metabarcoding approach targeting the ITS2 regions.Mycology. 2020 Aug 2;11(3):214-229. doi: 10.1080/21501203.2020.1799878. Mycology. 2020. PMID: 33062383 Free PMC article.
-
Taxonomy and systematics of plant probiotic bacteria in the genomic era.AIMS Microbiol. 2017 May 31;3(3):383-412. doi: 10.3934/microbiol.2017.3.383. eCollection 2017. AIMS Microbiol. 2017. PMID: 31294168 Free PMC article. Review.
-
Rediscovery of the microbial world in microbial ecology.Microbes Environ. 2013;28(3):281-4. doi: 10.1264/jsme2.me2803rh. Microbes Environ. 2013. PMID: 24064954 Free PMC article. No abstract available.
-
Genomic, biochemical, and phylogenetic evaluation of bacteria isolated from deep-sea sediment harboring methane hydrates.Arch Microbiol. 2022 Mar 9;204(4):205. doi: 10.1007/s00203-022-02814-z. Arch Microbiol. 2022. PMID: 35266047
-
Bacterial community structure and novel species of magnetotactic bacteria in sediments from a seamount in the Mariana volcanic arc.Sci Rep. 2017 Dec 21;7(1):17964. doi: 10.1038/s41598-017-17445-4. Sci Rep. 2017. PMID: 29269894 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ash C, Farrow JA, Dorsch M, Stackebrandt E, Collins MD. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. - PubMed
-
- Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–146. 148–149. - PubMed
-
- Benlloch S, López-López A, Casamayor EO, et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol. 2002;4:349–360. - PubMed
-
- Cardinale M, Brusetti L, Quatrini P, Borin S, Puglia AM, Rizzi A, Zanardini E, Sorlini C, Corselli C, Daffonchio D. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol. 2004;70:6147–6156. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

