"Epidemic clones" of Listeria monocytogenes are widespread and ancient clonal groups
- PMID: 24006010
- PMCID: PMC3889766
- DOI: 10.1128/JCM.01874-13
"Epidemic clones" of Listeria monocytogenes are widespread and ancient clonal groups
Abstract
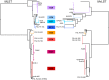
The food-borne pathogen Listeria monocytogenes is genetically heterogeneous. Although some clonal groups have been implicated in multiple outbreaks, there is currently no consensus on how "epidemic clones" should be defined. The objectives of this work were to compare the patterns of sequence diversity on two sets of genes that have been widely used to define L. monocytogenes clonal groups: multilocus sequence typing (MLST) and multi-virulence-locus sequence typing (MvLST). Further, we evaluated the diversity within clonal groups by pulsed-field gel electrophoresis (PFGE). Based on 125 isolates of diverse temporal, geographical, and source origins, MLST and MvLST genes (i) had similar patterns of sequence polymorphisms, recombination, and selection, (ii) provided concordant phylogenetic clustering, and (iii) had similar discriminatory power, which was not improved when we combined both data sets. Inclusion of representative strains of previous outbreaks demonstrated the correspondence of epidemic clones with previously recognized MLST clonal complexes. PFGE analysis demonstrated heterogeneity within major clones, most of which were isolated decades before their involvement in outbreaks. We conclude that the "epidemic clone" denominations represent a redundant but largely incomplete nomenclature system for MLST-defined clones, which must be regarded as successful genetic groups that are widely distributed across time and space.
Figures

References
-
- Denny J, McLauchlin J. 2008. Human Listeria monocytogenes infections in Europe: an opportunity for improved European surveillance. Euro Surveill. 13(13):pii=8082 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8082 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

