The TetR family of regulators
- PMID: 24006471
- PMCID: PMC3811609
- DOI: 10.1128/MMBR.00018-13
The TetR family of regulators
Abstract
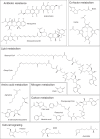
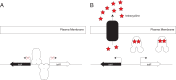
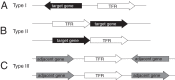
The most common prokaryotic signal transduction mechanisms are the one-component systems in which a single polypeptide contains both a sensory domain and a DNA-binding domain. Among the >20 classes of one-component systems, the TetR family of regulators (TFRs) are widely associated with antibiotic resistance and the regulation of genes encoding small-molecule exporters. However, TFRs play a much broader role, controlling genes involved in metabolism, antibiotic production, quorum sensing, and many other aspects of prokaryotic physiology. There are several well-established model systems for understanding these important proteins, and structural studies have begun to unveil the mechanisms by which they bind DNA and recognize small-molecule ligands. The sequences for more than 200,000 TFRs are available in the public databases, and genomics studies are identifying their target genes. Three-dimensional structures have been solved for close to 200 TFRs. Comparison of these structures reveals a common overall architecture of nine conserved α helices. The most important open question concerning TFR biology is the nature and diversity of their ligands and how these relate to the biochemical processes under their control.
Figures







References
-
- Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 - PubMed
-
- Schreiter ER, Drennan CL. 2007. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 5:710–720 - PubMed
-
- Pérez-Rueda E, Collado-Vides J. 2001. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J. Mol. Evol. 53:172–179 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

