Identification of upstream regulators for prognostic expression signature genes in colorectal cancer
- PMID: 24006872
- PMCID: PMC3847874
- DOI: 10.1186/1752-0509-7-86
Identification of upstream regulators for prognostic expression signature genes in colorectal cancer
Abstract
Background: Gene expression signatures have been commonly used as diagnostic and prognostic markers for cancer subtyping. However, expression signatures frequently include many passengers, which are not directly related to cancer progression. Their upstream regulators such as transcription factors (TFs) may take a more critical role as drivers or master regulators to provide better clues on the underlying regulatory mechanisms and therapeutic applications.
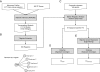
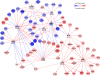
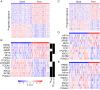
Results: In order to identify prognostic master regulators, we took the known 85 prognostic signature genes for colorectal cancer and inferred their upstream TFs. To this end, a global transcriptional regulatory network was constructed with total >200,000 TF-target links using the ARACNE algorithm. We selected the top 10 TFs as candidate master regulators to show the highest coverage of the signature genes among the total 846 TF-target sub-networks or regulons. The selected TFs showed a comparable or slightly better prognostic performance than the original 85 signature genes in spite of greatly reduced number of marker genes from 85 to 10. Notably, these TFs were selected solely from inferred regulatory links using gene expression profiles and included many TFs regulating tumorigenic processes such as proliferation, metastasis, and differentiation.
Conclusions: Our network approach leads to the identification of the upstream transcription factors for prognostic signature genes to provide leads to their regulatory mechanisms. We demonstrate that our approach could identify upstream biomarkers for a given set of signature genes with markedly smaller size and comparable performances. The utility of our method may be expandable to other types of signatures such as diagnosis and drug response.
Figures


References
-
- Méndez E, Lohavanichbutr P, Fan W, Houck JR, Rue TC, Doody DR, Futran ND, Upton MP, Yueh B, Zhao LP, Schwartz SM, Chen C. Can a metastatic gene expression profile outperform tumor size as a predictor of occult lymph node metastasis in oral cancer patients? Clin Cancer Res. 2011;17:2466–73. doi: 10.1158/1078-0432.CCR-10-0175. - DOI - PMC - PubMed
-
- Servant N, Bollet MA, Halfwerk H, Bleakley K, Kreike B, Jacob L, Sie D, Kerkhoven R, Hupe P, Hadhri R, Fourquet A, Bartelink H, Barillot E, Sigal-Zafrani B, Van De Vijver M. Search for a gene expression signature of breast cancer local recurrence in young women. Clin Cancer Res. 2012;45:1704–15. - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

