Splice variants of zinc finger protein 695 mRNA associated to ovarian cancer
- PMID: 24007497
- PMCID: PMC3847372
- DOI: 10.1186/1757-2215-6-61
Splice variants of zinc finger protein 695 mRNA associated to ovarian cancer
Abstract
Background: Studies of alternative mRNA splicing (AS) in health and disease have yet to yield the complete picture of protein diversity and its role in physiology and pathology. Some forms of cancer appear to be associated to certain alternative mRNA splice variants, but their role in the cancer development and outcome is unclear.
Methods: We examined AS profiles by means of whole genome exon expression microarrays (Affymetrix GeneChip 1.0) in ovarian tumors and ovarian cancer-derived cell lines, compared to healthy ovarian tissue. Alternatively spliced genes expressed predominantly in ovarian tumors and cell lines were confirmed by RT-PCR.
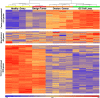
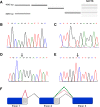
Results: Among several significantly overexpressed AS genes in malignant ovarian tumors and ovarian cancer cell lines, the most significant one was that of the zinc finger protein ZNF695, with two previously unknown mRNA splice variants identified in ovarian tumors and cell lines. The identity of ZNF695 AS variants was confirmed by cloning and sequencing of the amplicons obtained from ovarian cancer tissue and cell lines.
Conclusions: Alternative ZNF695 mRNA splicing could be a marker of ovarian cancer with possible implications on its pathogenesis.
Figures



Similar articles
-
Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20.Clin Cancer Res. 2004 Jul 15;10(14):4652-60. doi: 10.1158/1078-0432.CCR-03-0439. Clin Cancer Res. 2004. PMID: 15269137
-
Differentially expressed alternatively spliced genes in malignant pleural mesothelioma identified using massively parallel transcriptome sequencing.BMC Med Genet. 2009 Dec 31;10:149. doi: 10.1186/1471-2350-10-149. BMC Med Genet. 2009. PMID: 20043850 Free PMC article.
-
Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array.BMC Genomics. 2006 Dec 27;7:325. doi: 10.1186/1471-2164-7-325. BMC Genomics. 2006. PMID: 17192196 Free PMC article.
-
Splicing for alternative structures of Cav1.2 Ca2+ channels in cardiac and smooth muscles.Cardiovasc Res. 2005 Nov 1;68(2):197-203. doi: 10.1016/j.cardiores.2005.06.024. Epub 2005 Jul 27. Cardiovasc Res. 2005. PMID: 16051206 Review.
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
-
Gene Expression Profile of Cultured Human Coronary Arterial Endothelial Cells Exposed to Serum from Chronic Kidney Disease Patients: Role of MAPK Signaling Pathway.Int J Mol Sci. 2025 Apr 15;26(8):3732. doi: 10.3390/ijms26083732. Int J Mol Sci. 2025. PMID: 40332370 Free PMC article.
-
ZNF695, A Potential Prognostic Biomarker, Correlates with Im mune Infiltrates in Cervical Squamous Cell Carcinoma and Endoce rvical Adenocarcinoma: Bioinformatic Analysis and Experimental Verification.Curr Gene Ther. 2024;24(5):441-452. doi: 10.2174/0115665232285216240228071244. Curr Gene Ther. 2024. PMID: 38441026
-
Machine learning identification of molecular targets for medulloblastoma subgroups using microarray gene fingerprint analysis.Comput Struct Biotechnol J. 2025 Jul 24;27:3481-3491. doi: 10.1016/j.csbj.2025.07.033. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40808800 Free PMC article.
-
Promoter hypermethylation and comprehensive regulation of ncRNA lead to the down-regulation of ZNF880, providing a new insight for the therapeutics and research of colorectal cancer.BMC Med Genomics. 2023 Jun 27;16(1):148. doi: 10.1186/s12920-023-01571-2. BMC Med Genomics. 2023. PMID: 37370088 Free PMC article.
-
TFE3 and HIF1α regulates the expression of SHMT2 isoforms via alternative promoter utilization in ovarian cancer cells.Cell Death Dis. 2025 Mar 17;16(1):178. doi: 10.1038/s41419-025-07445-y. Cell Death Dis. 2025. PMID: 40097394 Free PMC article.
References
-
- Lancaster JM, Dressman HK, Clarke JP, Sayer RA, Martino MA, Cragun JM, Henriott AH, Gray J, Sutphen R, Elahi A. et al.Identification of genes associated with ovarian cancer metastasis using microarray expression analysis. Int J Gynecol Cancer. 2006;16:1733–1745. doi: 10.1111/j.1525-1438.2006.00660.x. - DOI - PubMed
-
- Maxwell GL, Chandramouli GV, Dainty L, Litzi TJ, Berchuck A, Barrett JC, Risinger JI. Microarray analysis of endometrial carcinomas and mixed mullerian tumors reveals distinct gene expression profiles associated with different histologic types of uterine cancer. Clin Cancer Res. 2005;11:4056–4066. doi: 10.1158/1078-0432.CCR-04-2001. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

