Pattern-recognition receptor signaling regulator mRNA expression in humans and mice, and in transient inflammation or progressive fibrosis
- PMID: 24009023
- PMCID: PMC3794773
- DOI: 10.3390/ijms140918124
Pattern-recognition receptor signaling regulator mRNA expression in humans and mice, and in transient inflammation or progressive fibrosis
Abstract
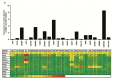
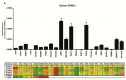
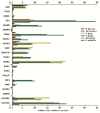
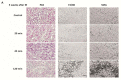
The cell type-, organ-, and species-specific expression of the pattern-recognition receptors (PRRs) are well described but little is known about the respective expression profiles of their negative regulators. We therefore determined the mRNA expression levels of A20, CYLD, DUBA, ST2, CD180, SIGIRR, TANK, SOCS1, SOCS3, SHIP, IRAK-M, DOK1, DOK2, SHP1, SHP2, TOLLIP, IRF4, SIKE, NLRX1, ERBIN, CENTB1, and Clec4a2 in human and mouse solid organs. Humans and mice displayed significant differences between their respective mRNA expression patterns of these factors. Additionally, we characterized their expression profiles in mononuclear blood cells upon bacterial endotoxin, which showed a consistent induction of A20, SOCS3, IRAK-M, and Clec4a2 in human and murine cells. Furthermore, we studied the expression pattern in transient kidney ischemia-reperfusion injury versus post-ischemic atrophy and fibrosis in mice. A20, CD180, ST2, SOCS1, SOCS3, SHIP, IRAK-M, DOK1, DOK2, IRF4, CENTB1, and Clec4a2 were all induced, albeit at different times of injury and repair. Progressive fibrosis was associated with a persistent induction of these factors. Thus, the organ- and species-specific expression patterns need to be considered in the design and interpretation of studies related to PRR-mediated innate immunity, which seems to be involved in tissue injury, tissue regeneration and in progressive tissue scarring.
Figures





Similar articles
-
Toll-like receptor and accessory molecule mRNA expression in humans and mice as well as in murine autoimmunity, transient inflammation, and progressive fibrosis.Int J Mol Sci. 2013 Jun 26;14(7):13213-30. doi: 10.3390/ijms140713213. Int J Mol Sci. 2013. PMID: 23803655 Free PMC article.
-
Quantitative expression of C-type lectin receptors in humans and mice.Int J Mol Sci. 2012;13(8):10113-10131. doi: 10.3390/ijms130810113. Epub 2012 Aug 14. Int J Mol Sci. 2012. PMID: 22949850 Free PMC article.
-
Differential role of Dok1 and Dok2 in TLR2-induced inflammatory signaling in glia.Mol Cell Neurosci. 2013 Sep;56:148-58. doi: 10.1016/j.mcn.2013.04.007. Epub 2013 May 7. Mol Cell Neurosci. 2013. PMID: 23659921
-
[Negative regulation of Toll-like receptor signalling].Postepy Hig Med Dosw (Online). 2013 Apr 25;67:339-50. doi: 10.5604/17322693.1046538. Postepy Hig Med Dosw (Online). 2013. PMID: 23619234 Review. Polish.
-
Intrinsic and extrinsic regulation of innate immune receptors.Yonsei Med J. 2011 May;52(3):379-92. doi: 10.3349/ymj.2011.52.3.379. Yonsei Med J. 2011. PMID: 21488180 Free PMC article. Review.
Cited by
-
At Least Two Genes Encode Many Variants of Irak3 in Rainbow Trout, but Neither the Full-Length Factor Nor Its Variants Interfere Directly With the TLR-Mediated Stimulation of Inflammation.Front Immunol. 2019 Sep 20;10:2246. doi: 10.3389/fimmu.2019.02246. eCollection 2019. Front Immunol. 2019. PMID: 31616422 Free PMC article.
-
Macrophage-Specific MCPIP1/Regnase-1 Attenuates Kidney Ischemia-Reperfusion Injury by Shaping the Local Inflammatory Response and Tissue Regeneration.Cells. 2022 Jan 24;11(3):397. doi: 10.3390/cells11030397. Cells. 2022. PMID: 35159206 Free PMC article.
-
Regulated necrosis-related molecule mRNA expression in humans and mice and in murine acute tissue injury and systemic autoimmunity leading to progressive organ damage, and progressive fibrosis.Biosci Rep. 2016 Dec 9;36(6):e00425. doi: 10.1042/BSR20160336. Print 2016 Dec. Biosci Rep. 2016. PMID: 27811014 Free PMC article.
-
Inflammation Regulation by Bacterial Molecular Patterns.Biomedicines. 2023 Jan 11;11(1):183. doi: 10.3390/biomedicines11010183. Biomedicines. 2023. PMID: 36672691 Free PMC article.
-
Immune system modulation of kidney regeneration--mechanisms and implications.Nat Rev Nephrol. 2014 Jun;10(6):347-58. doi: 10.1038/nrneph.2014.68. Epub 2014 Apr 29. Nat Rev Nephrol. 2014. PMID: 24776845 Review.
References
-
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

