The individualized genetic barrier predicts treatment response in a large cohort of HIV-1 infected patients
- PMID: 24009493
- PMCID: PMC3757085
- DOI: 10.1371/journal.pcbi.1003203
The individualized genetic barrier predicts treatment response in a large cohort of HIV-1 infected patients
Abstract
The success of combination antiretroviral therapy is limited by the evolutionary escape dynamics of HIV-1. We used Isotonic Conjunctive Bayesian Networks (I-CBNs), a class of probabilistic graphical models, to describe this process. We employed partial order constraints among viral resistance mutations, which give rise to a limited set of mutational pathways, and we modeled phenotypic drug resistance as monotonically increasing along any escape pathway. Using this model, the individualized genetic barrier (IGB) to each drug is derived as the probability of the virus not acquiring additional mutations that confer resistance. Drug-specific IGBs were combined to obtain the IGB to an entire regimen, which quantifies the virus' genetic potential for developing drug resistance under combination therapy. The IGB was tested as a predictor of therapeutic outcome using between 2,185 and 2,631 treatment change episodes of subtype B infected patients from the Swiss HIV Cohort Study Database, a large observational cohort. Using logistic regression, significant univariate predictors included most of the 18 drugs and single-drug IGBs, the IGB to the entire regimen, the expert rules-based genotypic susceptibility score (GSS), several individual mutations, and the peak viral load before treatment change. In the multivariate analysis, the only genotype-derived variables that remained significantly associated with virological success were GSS and, with 10-fold stronger association, IGB to regimen. When predicting suppression of viral load below 400 cps/ml, IGB outperformed GSS and also improved GSS-containing predictors significantly, but the difference was not significant for suppression below 50 cps/ml. Thus, the IGB to regimen is a novel data-derived predictor of treatment outcome that has potential to improve the interpretation of genotypic drug resistance tests.
Conflict of interest statement
I have read the journal's policy and have the following potential conflicts: HFG has been a medical adviser and/or consultant for GlaxoSmithKline, Abbott, Novartis, Boehringer Ingelheim, Gilead Sciences, Roche, Merck Sharp & Dohme, Tibotec, and Bristol-Myers Squibb, and has received unrestricted research, travel, and educational grants from Roche, Abbott, Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline, ViiV Healthcare, Tibotec and Merck Sharp & Dohme (all money sent to institution). SY has participated in advisory board of Bristol-Meyers Squibb, has received travel grants from ViiV and Merck Sharp & Dohme, and has been paid for development of educational presentations by Gilead. VvW was supported by a fellowship of the Novartis Foundation (formerly Ciba-Geigy Jubilee Foundation). HF's institution has received money from participation in advisory boards of ViiVHealthcare, Bristol-Myers Squibb, Gilead, Merck Sharp & Dome, Boehringer-Ingelheim, and Janssen, and has received unrestricted educational or research grants from Abbott, ViiV Healthcare, BMS, Roche, Gilead, Merck Sharp & Dome, and Janssen-Cilag. MB has been paid by ViiV, Gilead, and MSD for serving on advisory boards and his institution has received educational and research grants from ViiV, Boehringer, Gilead, Abbott, and Bristol-Meyers Squibb. MC has received travel grants from Abbott, Boehringer-Ingelheim, Gilead, and MSD. PV has been paid for consulting Bristol-Meyers Sqibb, Merck Sharp & Dohme, and Janssen, and for lecturing by Janssen and Gilead. EB has been paid by Boehringer Ingelheim, Gilead, Merck Sharp & Dohme, and ViiV for consultancy and board membership, and his institution has been paid by Janssen, Gilead, Abbott, Bristol-Meyers Squibb, and Merck Sharp & Dohme for board membership and consultancy.
Figures
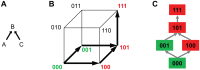
 ,
,  , and
, and  , is considered with the two relations
, is considered with the two relations  and
and  , resulting in two possible escape pathways of the virus, namely
, resulting in two possible escape pathways of the virus, namely  or
or  . (B) The partial order constraints give rise to the genotype lattice consisting of genotypes 000, 001, 100, 101, and 111 indicated with bold arrows, where genotypes are encoded as binary strings such that 000 is the wild type
. (B) The partial order constraints give rise to the genotype lattice consisting of genotypes 000, 001, 100, 101, and 111 indicated with bold arrows, where genotypes are encoded as binary strings such that 000 is the wild type  (no mutations), 100 is defined by mutation
(no mutations), 100 is defined by mutation  and identified with
and identified with  , 101 with
, 101 with  , etc. The genotype lattice
, etc. The genotype lattice  is shown inside the embedding hypercube
is shown inside the embedding hypercube  . For each antiretroviral drug, genotypes are labeled as either susceptible (green) or resistant (red). (C) Genotype lattice isolated from the embedding hypercube. The IGB is the probability of the virus not reaching a resistant state.
. For each antiretroviral drug, genotypes are labeled as either susceptible (green) or resistant (red). (C) Genotype lattice isolated from the embedding hypercube. The IGB is the probability of the virus not reaching a resistant state.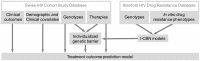
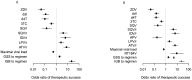
 ) and grey (
) and grey ( ) symbols. Only predictors with a p-value smaller than 0.01 are included.
) symbols. Only predictors with a p-value smaller than 0.01 are included.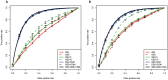
Similar articles
-
Does GSS still maintain relevance on HAART outcome after the introduction of newest active antiretroviral drugs? 48 weeks results.Curr HIV Res. 2011 Dec 1;9(8):625-9. doi: 10.2174/157016211798998790. Curr HIV Res. 2011. PMID: 22211659
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
-
Baseline HIV-1 resistance, virological outcomes, and emergent resistance in the SECOND-LINE trial: an exploratory analysis.Lancet HIV. 2015 Feb;2(2):e42-51. doi: 10.1016/S2352-3018(14)00061-7. Epub 2015 Jan 20. Lancet HIV. 2015. PMID: 26424460 Clinical Trial.
-
Improved prediction of response to antiretroviral combination therapy using the genetic barrier to drug resistance.Antivir Ther. 2007;12(2):169-78. doi: 10.1177/135965350701200202. Antivir Ther. 2007. PMID: 17503659
-
HIV-1 antiretroviral resistance: scientific principles and clinical applications.Drugs. 2012 Jun 18;72(9):e1-25. doi: 10.2165/11633630-000000000-00000. Drugs. 2012. PMID: 22686620 Free PMC article. Review.
Cited by
-
Nucleoside reverse-transcriptase inhibitor cross-resistance and outcomes from second-line antiretroviral therapy in the public health approach: an observational analysis within the randomised, open-label, EARNEST trial.Lancet HIV. 2017 Aug;4(8):e341-e348. doi: 10.1016/S2352-3018(17)30065-6. Epub 2017 May 8. Lancet HIV. 2017. PMID: 28495562 Free PMC article.
-
A framework for inferring fitness landscapes of patient-derived viruses using quasispecies theory.Genetics. 2015 Jan;199(1):191-203. doi: 10.1534/genetics.114.172312. Epub 2014 Nov 17. Genetics. 2015. PMID: 25406469 Free PMC article.
-
Decoding HIV resistance: from genotype to therapy.Future Med Chem. 2017 Sep;9(13):1529-1538. doi: 10.4155/fmc-2017-0048. Epub 2017 Aug 9. Future Med Chem. 2017. PMID: 28791894 Free PMC article. Review.
-
Lasso regularization for left-censored Gaussian outcome and high-dimensional predictors.BMC Med Res Methodol. 2018 Dec 4;18(1):159. doi: 10.1186/s12874-018-0609-4. BMC Med Res Methodol. 2018. PMID: 30514234 Free PMC article.
-
An Evolutionary Model-Based Approach To Quantify the Genetic Barrier to Drug Resistance in Fast-Evolving Viruses and Its Application to HIV-1 Subtypes and Integrase Inhibitors.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00539-19. doi: 10.1128/AAC.00539-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31109980 Free PMC article.
References
-
- Thompson MA, Aberg JA, Cahn P, Montaner JSG, Rizzardini G, et al. (2010) Antiretroviral treatment of adult HIV infection: 2010 recommendations of the international AIDS Society-USA panel. JAMA 304: 321–333. - PubMed
-
- Hirsch MS, Günthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, et al. (2008) Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an international AIDS Society-USA panel. Clin Infect Dis 47: 266–285. - PubMed
-
- Saigo H, Altmann A, Bogojeska J, Mueller F, Nowozin S, et al. (2011) Learning from past treatments and their outcome improves prediction of in vivo response to anti-HIV therapy. Stat Appl Genet Mol Biol 10: Article 6. - PubMed
-
- Jiang H, Deeks SG, Kuritzkes DR, Lallemant M, Katzenstein D, et al. (2003) Assessing resistance costs of antiretroviral therapies via measures of future drug options. J Infect Dis 188: 1001–1008. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

