Influenza a virus migration and persistence in North American wild birds
- PMID: 24009503
- PMCID: PMC3757048
- DOI: 10.1371/journal.ppat.1003570
Influenza a virus migration and persistence in North American wild birds
Abstract
Wild birds have been implicated in the emergence of human and livestock influenza. The successful prediction of viral spread and disease emergence, as well as formulation of preparedness plans have been hampered by a critical lack of knowledge of viral movements between different host populations. The patterns of viral spread and subsequent risk posed by wild bird viruses therefore remain unpredictable. Here we analyze genomic data, including 287 newly sequenced avian influenza A virus (AIV) samples isolated over a 34-year period of continuous systematic surveillance of North American migratory birds. We use a Bayesian statistical framework to test hypotheses of viral migration, population structure and patterns of genetic reassortment. Our results reveal that despite the high prevalence of Charadriiformes infected in Delaware Bay this host population does not appear to significantly contribute to the North American AIV diversity sampled in Anseriformes. In contrast, influenza viruses sampled from Anseriformes in Alberta are representative of the AIV diversity circulating in North American Anseriformes. While AIV may be restricted to specific migratory flyways over short time frames, our large-scale analysis showed that the long-term persistence of AIV was independent of bird flyways with migration between populations throughout North America. Analysis of long-term surveillance data provides vital insights to develop appropriately informed predictive models critical for pandemic preparedness and livestock protection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

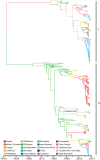
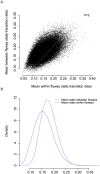
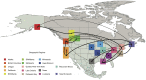
References
-
- Chen H, Smith GJD, Zhang SY, Qin K, Wang J, et al. (2005) Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436: 191–192. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

