Variable Expression of GLIPR1 Correlates with Invasive Potential in Melanoma Cells
- PMID: 24010123
- PMCID: PMC3757444
- DOI: 10.3389/fonc.2013.00225
Variable Expression of GLIPR1 Correlates with Invasive Potential in Melanoma Cells
Abstract
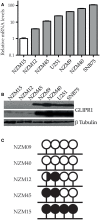
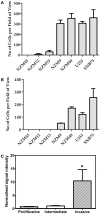
GLI pathogenesis-related 1 (GLIPR1) was previously identified as an epigenetically regulated tumor suppressor in prostate cancer and, conversely, an oncoprotein in glioma. More recently, GLIPR1 was shown to be differentially expressed in other cancers including ovarian, acute myeloid leukemia, and Wilms' tumor. Here we investigated GLIPR1 expression in metastatic melanoma cell lines and tissue. GLIPR1 was variably expressed in metastatic melanoma cells, and transcript levels correlated with degree of GLIPR1 promoter methylation in vitro. Elevated GLIPR1 levels were correlated with increased invasive potential, and siRNA-mediated knockdown of GLIPR1 expression resulted in reduced cell migration and proliferation in vitro. Immunohistochemical studies of melanoma tissue microarrays showed moderate to high staining for GLIPR1 in 50% of specimens analyzed. GLIPR1 staining was observed in normal skin in merocrine sweat glands, sebaceous glands, and hair follicles within the dermis.
Keywords: CAP; GLIPR1; invasion; melanoma; methylation.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

