Allosteric conformational barcodes direct signaling in the cell
- PMID: 24010710
- PMCID: PMC6361540
- DOI: 10.1016/j.str.2013.06.002
Allosteric conformational barcodes direct signaling in the cell
Abstract
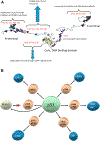
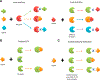
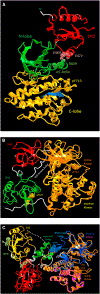
The cellular network is highly interconnected. Pathways merge and diverge. They proceed through shared proteins and may change directions. How are cellular pathways controlled and their directions decided, coded, and read? These questions become particularly acute when we consider that a small number of pathways, such as signaling pathways that regulate cell fates, cell proliferation, and cell death in development, are extensively exploited. This review focuses on these signaling questions from the structural standpoint and discusses the literature in this light. All co-occurring allosteric events (including posttranslational modifications, pathogen binding, and gain-of-function mutations) collectively tag the protein functional site with a unique barcode. The barcode shape is read by an interacting molecule, which transmits the signal. A conformational barcode provides an intracellular address label, which selectively favors binding to one partner and quenches binding to others, and, in this way, determines the pathway direction, and, eventually, the cell's response and fate.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Antal MA, Bode C, and Csermely P (2009). Perturbation waves in proteins and protein networks: applications of percolation and game theories in signaling and drug design. Curr. Protein Pept. Sci. 10, 161–172. - PubMed
-
- Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, Mizushima T, Ichinose K, Nakamura H, Tsujino A, Kawakami A,et al. (2011). Protea- some assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. USA 108, 14914–14919. - PMC - PubMed
-
- Benayoun BA, and Veitia RA (2009). A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol. 19, 189–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

