Pseudo attP sites in favor of transgene integration and expression in cultured porcine cells identified by Streptomyces phage phiC31 integrase
- PMID: 24010979
- PMCID: PMC3844521
- DOI: 10.1186/1471-2199-14-20
Pseudo attP sites in favor of transgene integration and expression in cultured porcine cells identified by Streptomyces phage phiC31 integrase
Abstract
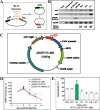
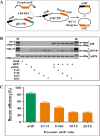
Phage PhiC31 integrase integrates attB-containing plasmid into pseudo attP site in eukaryotic genomes in a unidirectional site-specific manner and maintains robust transgene expression. Few studies, however, explore its potential in livestock. This study aims to discover the molecular basis of PhiC31 integrase-mediated site-specific recombination in pig cells. We show that PhiC31 integrase can mediate site-specific transgene integration into the genome of pig kidney PK15 cells. Intramolecular recombination in pig PK15 cell line occurred at maximum frequency of 82% with transiently transfected attB- and attP-containing plasmids. An optimal molar ratio of pCMV-Int to pEGFP-N1-attB at 5:1 was observed for maximum number of cell clones under drug selection. Four candidate pseudo attP sites were identified by TAIL-PCR from those cell clones with single-copy transgene integration. Two of them gave rise to higher integration frequency occurred at 33%. 5' and 3' junction PCR showed that transgene integration mediated by PhiC31 integrase was mono-allelic. Micro- deletion and insertion were observed by sequencing the integration border, indicating that double strand break was induced by the recombination. We then constructed rescue reporter plasmids by ABI-REC cloning of the four pseudo attP sites into pBCPB + plasmid. Transfection of these rescue plasmids and pCMV-Int resulted in expected intramolecular recombination between attB and pseudo attP sites. This proved that the endogenous pseudo attP sites were functional substrates for PhiC31 integrase-mediated site-specific recombination. Two pseudo attP sites maintained robust extracellular and intracellular EGFP expression. Alamar blue assay showed that transgene integration into these specific sites had little effect on cell proliferation. This is the first report to document the potential use of PhiC31 integrase to mediate site-specific recombination in pig cells. Our work established an ideal model to study the position effect of identical transgene located in diverse chromosomal contexts. These findings also form the basis for targeted pig genome engineering and may be used to produce genetically modified pigs for agricultural and biomedical uses.
Figures



References
-
- Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, Schmidt M, Winther KD, Schyth BD, Holm IE, Vajta G, Bolund L, Callesen H, Jørgensen AL, Nielsen AL, Mikkelsen JG. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res. 2011;20:533–545. doi: 10.1007/s11248-010-9438-x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

