Engineered fungal polyketide biosynthesis in Pichia pastoris: a potential excellent host for polyketide production
- PMID: 24011431
- PMCID: PMC3847973
- DOI: 10.1186/1475-2859-12-77
Engineered fungal polyketide biosynthesis in Pichia pastoris: a potential excellent host for polyketide production
Abstract
Background: Polyketides are one of the most important classes of secondary metabolites and usually make good drugs. Currently, heterologous production of fungal polyketides for developing a high potential industrial application system with high production capacity and pharmaceutical feasibility was still at its infancy. Pichia pastoris is a highly successful system for the high production of a variety of heterologous proteins. In this work, we aim to develop a P. pastoris based in vivo fungal polyketide production system for first time and evaluate its feasibility for future industrial application.
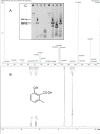
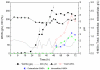
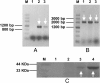
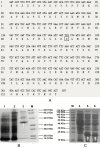
Results: A recombinant P. pastoris GS115-NpgA-ATX with Aspergillus nidulans phosphopantetheinyl transferase (PPtase) gene npgA and Aspergillus terrus 6-methylsalicylic acid (6-MSA) synthase (6-MSAS) gene atX was constructed. A specific compound was isolated and identified as 6-MSA by HPLC, LC-MS and NMR. Transcription of both genes were detected. In 5-L bioreactor, the GS115-NpgA-ATX grew well and produced 6-MSA quickly until reached a high value of 2.2 g/L by methanol induction for 20 hours. Thereafter, the cells turned to death ascribing to high concentration of antimicrobial 6-MSA. The distribution of 6-MSA changed that during early and late induction phase it existed more in supernatant while during intermediate stage it mainly located intracellular. Different from 6-MSA production strain, recombinant M. purpureus pksCT expression strains for citrinin intermediate production, no matter PksCT located in cytoplasm or in peroxisomes, did not produce any specific compound. However, both npgA and pksCT transcripted effectively in cells and western blot analysis proved the expression of PPtase. Then the PPTase was expressed and purified, marked by fluorescent probes, and reacted with purified ACP domain and its mutant ACPm of PksCT. Fluoresence was only observed in ACP but not ACPm, indicating that the PPTase worked well with ACP to make it bioactive holo-ACP. Thus, some other factors may affect polyketide synthesis that include activities of the individual catalytic domains and release of the product from the synthase of PksCT.
Conclusions: An efficient P. pastoris expression system of fungal polyketides was successfully constructed. It produced a high production of 6-MSA and holds potential for future industrial application of 6-MSA and other fungal polyketides.
Figures


Similar articles
-
Methylotrophic yeast Pichia pastoris as a chassis organism for polyketide synthesis via the full citrinin biosynthetic pathway.J Biotechnol. 2017 Jan 20;242:64-72. doi: 10.1016/j.jbiotec.2016.11.031. Epub 2016 Nov 29. J Biotechnol. 2017. PMID: 27913218
-
De novo production of aromatic m-cresol in Saccharomyces cerevisiae mediated by heterologous polyketide synthases combined with a 6-methylsalicylic acid decarboxylase.Metab Eng Commun. 2019 May 4;9:e00093. doi: 10.1016/j.mec.2019.e00093. eCollection 2019 Dec. Metab Eng Commun. 2019. PMID: 31193192 Free PMC article.
-
Optimization of heterologous production of the polyketide 6-MSA in Saccharomyces cerevisiae.Biotechnol Bioeng. 2007 Jul 1;97(4):893-900. doi: 10.1002/bit.21286. Biotechnol Bioeng. 2007. PMID: 17171715
-
Heterologous protein expression in the methylotrophic yeast Pichia pastoris.FEMS Microbiol Rev. 2000 Jan;24(1):45-66. doi: 10.1111/j.1574-6976.2000.tb00532.x. FEMS Microbiol Rev. 2000. PMID: 10640598 Review.
-
Heterologous protein production using the Pichia pastoris expression system.Yeast. 2005 Mar;22(4):249-70. doi: 10.1002/yea.1208. Yeast. 2005. PMID: 15704221 Review.
Cited by
-
Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris.Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2201711119. doi: 10.1073/pnas.2201711119. Epub 2022 Jul 11. Proc Natl Acad Sci U S A. 2022. PMID: 35858340 Free PMC article.
-
A hybrid pathway for self-sustained luminescence.Sci Adv. 2024 Mar 8;10(10):eadk1992. doi: 10.1126/sciadv.adk1992. Epub 2024 Mar 8. Sci Adv. 2024. PMID: 38457503 Free PMC article.
-
Engineering the synthetic β-alanine pathway in Komagataella phaffii for conversion of methanol into 3-hydroxypropionic acid.Microb Cell Fact. 2023 Nov 17;22(1):237. doi: 10.1186/s12934-023-02241-9. Microb Cell Fact. 2023. PMID: 37978380 Free PMC article.
-
Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis.Biomolecules. 2020 Jun 6;10(6):874. doi: 10.3390/biom10060874. Biomolecules. 2020. PMID: 32517243 Free PMC article. Review.
-
Pathway reconstruction and metabolic engineering for the de novo and enhancing production of monacolin J in Pichia pastoris.Bioprocess Biosyst Eng. 2024 Nov;47(11):1789-1801. doi: 10.1007/s00449-024-03069-2. Epub 2024 Jul 31. Bioprocess Biosyst Eng. 2024. PMID: 39085651
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

