Epigenetic modifications in the pathogenesis of diabetic nephropathy
- PMID: 24011576
- PMCID: PMC3767931
- DOI: 10.1016/j.semnephrol.2013.05.006
Epigenetic modifications in the pathogenesis of diabetic nephropathy
Abstract
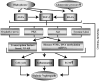
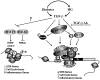
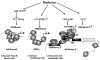
Diabetic nephropathy (DN) is a leading cause of end-stage renal disease. Diabetic vascular complications such as DN can progress despite subsequent glycemic control, suggesting a metabolic memory of previous exposure to hyperglycemia. Diabetes profoundly impacts transcription programs in target cells through activation of multiple signaling pathways and key transcription factors leading to aberrant expression of pathologic genes. Emerging evidence suggests that these factors associated with the pathophysiology of diabetic complications and metabolic memory also might be influenced by epigenetic mechanisms in chromatin such as DNA methylation, histone lysine acetylation, and methylation. Key histone modifications and the related histone methyltransferases and acetyltransferases have been implicated in the regulation of inflammatory and profibrotic genes in renal and vascular cells under diabetic conditions. Advances in epigenome profiling approaches have provided novel insights into the chromatin states and functional outcomes in target cells affected by diabetes. Because epigenetic changes are potentially reversible, they can provide a window of opportunity for the development of much-needed new therapies for DN in the future. In this review, we discuss recent developments in the field of epigenetics and their relevance to diabetic vascular complications and DN pathogenesis.
Keywords: Chromatin; diabetic nephropathy; epigenomics; histone modifications; metabolic memory.
© 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
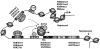
References
-
- Sanchez AP, Sharma K. Transcription factors in the pathogenesis of diabetic nephropathy. Expert Rev Mol Med. 2009;11:e13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

