Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study
- PMID: 24011653
- PMCID: PMC3879782
- DOI: 10.1016/S1474-4422(13)70210-2
Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study
Abstract
Background: Hexanucleotide repeat expansions in chromosome 9 open reading frame 72 (C9ORF72) are the most common known genetic cause of frontotemporal dementia (FTD) and motor neuron disease (MND). We assessed whether expansion size is associated with disease severity or phenotype.
Methods: We did a cross-sectional Southern blot characterisation study (Xpansize-72) in a cohort of individuals with FTD, MND, both these diseases, or no clinical phenotype. All participants had GGGGCC repeat expansions in C9ORF72, and high quality DNA was available from one or more of the frontal cortex, cerebellum, or blood. We used Southern blotting techniques and densitometry to estimate the repeat size of the most abundant expansion species. We compared repeat sizes between different tissues using Wilcoxon rank sum and Wilcoxon signed rank tests, and between disease subgroups using Kruskal-Wallis rank sum tests. We assessed the association of repeat size with age at onset and age at collection using a Spearman's test of correlation, and assessed the association between repeat size and survival after disease onset using Cox proportional hazards regression models.
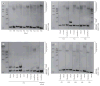
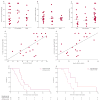
Findings: We included 84 individuals with C9ORF72 expansions: 35 had FTD, 16 had FTD and MND, 30 had MND, and three had no clinical phenotype. We focused our analysis on three major tissue subgroups: frontal cortex (available from 41 patients [21 with FTD, 11 with FTD and MND, and nine with MND]), cerebellum (40 patients [20 with FTD, 12 with FTD and MND, and eight with MND]), and blood (47 patients [15 with FTD, nine with FTD and MND, and 23 with MND] and three carriers who had no clinical phenotype). Repeat lengths in the cerebellum were smaller (median 12·3 kb [about 1667 repeat units], IQR 11·1-14·3) than those in the frontal cortex (33·8 kb [about 5250 repeat units], 23·5-44·9; p<0·0001) and those in blood (18·6 kb [about 2717 repeat units], 13·9-28·1; p=0·0002). Within these tissues, we detected no difference in repeat length between disease subgroups (cerebellum p=0·96, frontal cortex p=0·27, blood p=0·10). In the frontal cortex of patients with FTD, repeat length correlated with age at onset (r=0·63; p=0·003) and age at sample collection (r=0·58; p=0·006); we did not detect such a correlation in samples from the cerebellum or blood. When assessing cerebellum samples from the overall cohort, survival after disease onset was 4·8 years (IQR 3·0-7·4) in the group with expansions greater than 1467 repeat units (the 25th percentile of repeat lengths) versus 7·4 years (6·3-10·9) in the group with smaller expansions (HR 3·27, 95% CI 1·34-7·95; p=0·009).
Interpretation: We detected substantial variation in repeat sizes between samples from the cerebellum, frontal cortex, and blood, and longer repeat sizes in the cerebellum seem to be associated with a survival disadvantage. Our findings indicate that expansion size does affect disease severity, which--if replicated in other cohorts--could be relevant for genetic counselling.
Funding: The ALS Therapy Alliance, the National Institute of Neurological Disorders and Stroke, the National Institute on Aging, the Arizona Department of Health Services, the Arizona Biomedical Research Commission, and the Michael J Fox Foundation for Parkinson's Research.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Conflict of interest statement
MDJ and RR hold a patent on methods to screen for the hexanucleotide repeat expansion in the
Figures

Comment in
-
ALS and FTD: two sides of the same coin?Lancet Neurol. 2013 Oct;12(10):937-8. doi: 10.1016/S1474-4422(13)70218-7. Epub 2013 Sep 5. Lancet Neurol. 2013. PMID: 24011654 No abstract available.
References
-
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

