The EBAX-type Cullin-RING E3 ligase and Hsp90 guard the protein quality of the SAX-3/Robo receptor in developing neurons
- PMID: 24012004
- PMCID: PMC3779136
- DOI: 10.1016/j.neuron.2013.06.035
The EBAX-type Cullin-RING E3 ligase and Hsp90 guard the protein quality of the SAX-3/Robo receptor in developing neurons
Abstract
Although protein quality control (PQC) is generally perceived as important for the development of the nervous system, the specific mechanisms of neuronal PQC have remained poorly understood. Here, we report that C. elegans Elongin BC-binding axon regulator (EBAX-1), a conserved BC-box protein, regulates axon guidance through PQC of the SAX-3/Robo receptor. EBAX-1 buffers guidance errors against temperature variations. As a substrate-recognition subunit in the Elongin BC-containing Cullin-RING ubiquitin ligase (CRL), EBAX-1 also binds to DAF-21, a cytosolic Hsp90 chaperone. The EBAX-type CRL and DAF-21 collaboratively regulate SAX-3-mediated axon pathfinding. Biochemical and imaging assays indicate that EBAX-1 specifically recognizes misfolded SAX-3 and promotes its degradation in vitro and in vivo. Importantly, vertebrate EBAX also shows substrate preference toward aberrant Robo3 implicated in horizontal gaze palsy with progressive scoliosis (HGPPS). Together, our findings demonstrate a triage PQC mechanism mediated by the EBAX-type CRL and DAF-21/Hsp90 that maintains the accuracy of neuronal wiring.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
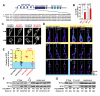
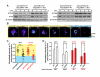
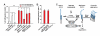
References
-
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. - PubMed
-
- Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

