Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2
- PMID: 24012420
- PMCID: PMC3831370
- DOI: 10.1016/j.immuni.2013.08.025
Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2
Abstract
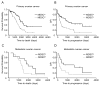
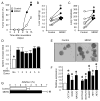
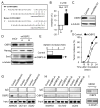
Myeloid-derived suppressor cells (MDSCs) and cancer stem cells (CSCs) are important cellular components in the cancer microenvironment and may affect cancer phenotype and patient outcome. The nature of MDSCs and their interaction with CSCs in ovarian carcinoma are unclear. We examined the interaction between MDSCs and CSCs in patients with ovarian carcinoma and showed that MDSCs inhibited T cell activation and enhanced CSC gene expression, sphere formation, and cancer metastasis. MDSCs triggered miRNA101 expression in cancer cells. miRNA101 subsequently repressesed the corepressor gene C-terminal binding protein-2 (CtBP2), and CtBP2 directly targeted stem cell core genes resulting in increased cancer cell stemness and increasing metastatic and tumorigenic potential. Increased MDSC density and tumor microRNA101 expression predict poor survival, as does decreased tumor CtBP2 expression, independent of each other. Collectively, our work identifies an immune-associated cellular, molecular, and clinical network involving MDSCs-microRNA101-CtBP2-stem cell core genes, which extrinsically controls cancer stemness and impacts patient outcome.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

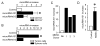


References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. - PubMed
-
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. - PubMed
-
- Cui TX, Kwok R, Schwartz J. Cooperative regulation of endogenous cAMP-response element binding protein and CCAAT/enhancer-binding protein beta in GH-stimulated c-fos expression. J Endocrinol. 2008;196:89–100. - PubMed
-
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

