Epigenetic changes in the progression of Alzheimer's disease
- PMID: 24012631
- PMCID: PMC3857018
- DOI: 10.1016/j.mad.2013.08.005
Epigenetic changes in the progression of Alzheimer's disease
Abstract
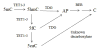
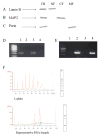
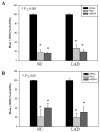
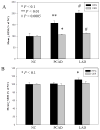
The formation of 5-hydroxymethylcytosine (5hmC), a key intermediate of DNA demethylation, is driven by the ten eleven translocation (TET) family of proteins that oxidize 5-methylcytosine (5mC) to 5hmC. To determine whether methylation/demethylation status is altered during the progression of Alzheimer's disease (AD), levels of TET1, 5mC and subsequent intermediates, including 5hmC, 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) were quantified in nuclear DNA from the hippocampus/parahippocampal gyrus (HPG) and the cerebellum of 5 age-matched normal controls, 5 subjects with preclinical AD (PCAD) and 7 late-stage AD (LAD) subjects by immunochemistry. The results showed significantly (p < 0.05) increased levels of TET1, 5mC, and 5hmC in the HPG of PCAD and LAD subjects. In contrast, levels of 5fC and 5caC were significantly (p < 0.05) decreased in the HPG of PCAD and LAD subjects. Overall, the data suggest altered methylation/demethylation patterns in vulnerable brain regions prior to the onset of clinical symptoms in AD suggesting a role in the pathogenesis of the disease.
Keywords: 5-Hydroxymethylcytosine; 5-Methylcytosine; Alzheimer's disease; Preclinical Alzheimer's disease.
Copyright © 2013 Elsevier Ireland Ltd. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

