A panel of 16 full-length HCV genomes was characterized in China belonging to genotypes 1-6 including subtype 2f and two novel genotype 6 variants
- PMID: 24012950
- PMCID: PMC4186718
- DOI: 10.1016/j.meegid.2013.08.014
A panel of 16 full-length HCV genomes was characterized in China belonging to genotypes 1-6 including subtype 2f and two novel genotype 6 variants
Abstract
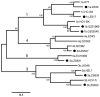
In this study, we characterized the full-length genomes of 16 HCV isolates obtained from patients in a single hospital in China using overlapping PCR followed by DNA sequencing. The obtained genomes are 9414-9628 nucleotides in length, and each genome contains a single ORF of 9021-9102 nucleotides. Nine genomes represent the common subtypes 1a, 1b, 2a, 2b, 3a, 3b, and 6a, while seven represent the infrequent lineages 1c, 2f, 4d, and 5a, and two novel genotype 6 variants. GZ51969 and GZ52540 are subtype 1b isolates belonging to two unique clusters designated A and B, which account for 29.5% and 59.5% of the 1b infections in China, respectively. ZS542 and GZ98799 represent the first two complete genomes of the provisionally assigned subtype 2f. ZS96 and ZS202 are novel genotype 6 variants that may qualify for two new subtypes. ZS17, ZS537, and ZS631 represent three alien subtypes, namely, 1c, 4d, and 5a, which were detected in China for the first time in this study and may have been recently introduced as a result of globalization. Taken together, these results confirmed a large variety of HCV taxonomic lineages in China through the sequencing of their full-length genomes. These lineages represent six genotypes, 11 subtypes, and two novel variants. They were characterized for achieving a better understanding of the HCV genetic variation patterns and for possible future research applications.
Keywords: China; Full-length genome; Genotype; HCV; Subtype.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
An increased diversity of HCV isolates were characterized among 393 patients with liver disease in China representing six genotypes, 12 subtypes, and two novel genotype 6 variants.J Clin Virol. 2013 Aug;57(4):311-7. doi: 10.1016/j.jcv.2013.04.013. Epub 2013 May 21. J Clin Virol. 2013. PMID: 23706765 Free PMC article.
-
Full-length genomes of 16 hepatitis C virus genotype 1 isolates representing subtypes 1c, 1d, 1e, 1g, 1h, 1i, 1j and 1k, and two new subtypes 1m and 1n, and four unclassified variants reveal ancestral relationships among subtypes.J Gen Virol. 2014 Jul;95(Pt 7):1479-1487. doi: 10.1099/vir.0.064980-0. Epub 2014 Apr 9. J Gen Virol. 2014. PMID: 24718832 Free PMC article.
-
Analysis of HCV Isolates Among the Li Ethnic in Hainan Island of South China Reveals Their HCV-6 Unique Evolution and a New Subtype.Cell Physiol Biochem. 2018;50(5):1832-1839. doi: 10.1159/000494863. Epub 2018 Nov 5. Cell Physiol Biochem. 2018. PMID: 30396187
-
An expanded taxonomy of hepatitis C virus genotype 6: Characterization of 22 new full-length viral genomes.Virology. 2015 Feb;476:355-363. doi: 10.1016/j.virol.2014.12.025. Epub 2015 Jan 10. Virology. 2015. PMID: 25589238 Free PMC article.
-
A new HCV genotype 6 subtype designated 6v was confirmed with three complete genome sequences.J Clin Virol. 2009 Mar;44(3):195-9. doi: 10.1016/j.jcv.2008.12.009. J Clin Virol. 2009. PMID: 19179105
Cited by
-
The distribution of hepatitis C viral genotypes shifted among chronic hepatitis C patients in Yunnan, China, between 2008-2018.Front Cell Infect Microbiol. 2023 Jul 11;13:1092936. doi: 10.3389/fcimb.2023.1092936. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37496804 Free PMC article.
-
Migration patterns of hepatitis C virus in China characterized for five major subtypes based on samples from 411 volunteer blood donors from 17 provinces and municipalities.J Virol. 2014 Jul;88(13):7120-9. doi: 10.1128/JVI.00414-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719413 Free PMC article.
-
Origin of hepatitis C virus genotype 3 in Africa as estimated through an evolutionary analysis of the full-length genomes of nine subtypes, including the newly sequenced 3d and 3e.J Gen Virol. 2014 Aug;95(Pt 8):1677-1688. doi: 10.1099/vir.0.065128-0. Epub 2014 May 2. J Gen Virol. 2014. PMID: 24795446 Free PMC article.
-
Genotype Distribution and Molecular Epidemiology of Hepatitis C Virus in Hubei, Central China.PLoS One. 2015 Sep 1;10(9):e0137059. doi: 10.1371/journal.pone.0137059. eCollection 2015. PLoS One. 2015. PMID: 26325070 Free PMC article.
-
The evolutionary dynamics and epidemiological history of hepatitis C virus genotype 6, including unique strains from the Li community of Hainan Island, China.Virus Evol. 2022 Feb 16;8(1):veac012. doi: 10.1093/ve/veac012. eCollection 2022. Virus Evol. 2022. PMID: 35600095 Free PMC article.
References
-
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

