Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma
- PMID: 24012984
- PMCID: PMC3892430
- DOI: 10.1053/j.gastro.2013.08.055
Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma
Abstract
Background & aims: Epigenetic silencing of tumor suppressor genes contributes to the pathogenesis of hepatocellular carcinoma (HCC). To identify clinically relevant tumor suppressor genes silenced by DNA methylation in HCC, we integrated DNA methylation data from human primary HCC samples with data on up-regulation of gene expression after epigenetic unmasking.
Methods: We performed genome-wide methylation analysis of 71 human HCC samples using the Illumina HumanBeadchip27K array; data were combined with those from microarray analysis of gene re-expression in 4 liver cancer cell lines after their exposure to reagents that reverse DNA methylation (epigenetic unmasking).
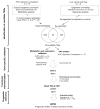
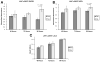
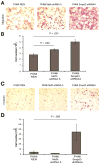
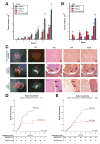
Results: Based on DNA methylation in primary HCC and gene re-expression in cell lines after epigenetic unmasking, we identified 13 candidate tumor suppressor genes. Subsequent validation led us to focus on functionally characterizing 2 candidates, sphingomyelin phosphodiesterase 3 (SMPD3) and neurofilament, heavy polypeptide (NEFH), which we found to behave as tumor suppressor genes in HCC. Overexpression of SMPD3 and NEFH by stable transfection of inducible constructs into an HCC cell line reduced cell proliferation by 50% and 20%, respectively (SMPD3, P = .003 and NEFH, P = .003). Conversely, knocking down expression of these genes with small hairpin RNA promoted cell invasion and migration in vitro (SMPD3, P = .0001 and NEFH, P = .022), and increased their ability to form tumors after subcutaneous injection or orthotopic transplantation into mice, confirming their role as tumor suppressor genes in HCC. Low levels of SMPD3 were associated with early recurrence of HCC after curative surgery in an independent patient cohort (P = .001; hazard ratio = 3.22; 95% confidence interval: 1.6-6.5 in multivariate analysis).
Conclusions: Integrative genomic analysis identified SMPD3 and NEFH as tumor suppressor genes in HCC. We provide evidence that SMPD3 is a potent tumor suppressor gene that could affect tumor aggressiveness; a reduced level of SMPD3 is an independent prognostic factor for early recurrence of HCC.
Keywords: 5-aza-2-deoxycitidine; 5-aza-2′deoxycitidine; ACTL6B; CHTN; CpG; DAC; DGKI; ELOVL fatty acid elongase 4; ELOVL4; GSTP1; HCC; LDHB; LRAT; ND; NEFH; PRPH; SMPD3; Sphingomyelin Phosphodiesterase; TSA; actin-like 6B; cooperative tissue network; cytosine preceding guanine; diacylglycerol kinase, iota; glutathione S-transferase 1; hepatocellular carcinoma; lactate dehydrogenase B; lecithin retinol acyltransferase (phosphatidylcholine−retinol O-acyltransferase); nSMase2; neurofilament, heavy polypeptide; nondiseased; peripherin; shRNA; short hairpin RNA; sphingomyelin phosphodiesterase 3; trichostatin A.
Copyright © 2013 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest
The authors disclose no conflicts.
Figures

References
-
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. - PubMed
-
- Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10:34–42. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

