Metabolic imaging in multiple time scales
- PMID: 24013043
- PMCID: PMC3942365
- DOI: 10.1016/j.ymeth.2013.08.027
Metabolic imaging in multiple time scales
Abstract
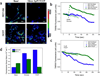
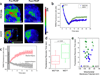
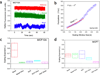
We report here a novel combination of time-resolved imaging methods for probing mitochondrial metabolism in multiple time scales at the level of single cells. By exploiting a mitochondrial membrane potential reporter fluorescence we demonstrate the single cell metabolic dynamics in time scales ranging from microseconds to seconds to minutes in response to glucose metabolism and mitochondrial perturbations in real time. Our results show that in comparison with normal human mammary epithelial cells, the breast cancer cells display significant alterations in metabolic responses at all measured time scales by single cell kinetics, fluorescence recovery after photobleaching and by scaling analysis of time-series data obtained from mitochondrial fluorescence fluctuations. Furthermore scaling analysis of time-series data in living cells with distinct mitochondrial dysfunction also revealed significant metabolic differences thereby suggesting the broader applicability (e.g. in mitochondrial myopathies and other metabolic disorders) of the proposed strategies beyond the scope of cancer metabolism. We discuss the scope of these findings in the context of developing portable, real-time metabolic measurement systems that can find applications in preclinical and clinical diagnostics.
Keywords: Breast cancer; Metabolic imaging; Microscopy; Mitochondria; NDUFS3; Scaling behavior.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Single-cell time-lapse imaging of intracellular O2 in response to metabolic inhibition and mitochondrial cytochrome-c release.Cell Death Dis. 2017 Jun 1;8(6):e2853. doi: 10.1038/cddis.2017.247. Cell Death Dis. 2017. PMID: 28569778 Free PMC article.
-
A faster, high resolution, mtPA-GFP-based mitochondrial fusion assay acquiring kinetic data of multiple cells in parallel using confocal microscopy.J Vis Exp. 2012 Jul 20;(65):e3991. doi: 10.3791/3991. J Vis Exp. 2012. PMID: 22847388 Free PMC article.
-
Fluorescence Probing Live Single-cell Mass Spectrometry for Direct Analysis of Organelle Metabolism.Anal Sci. 2015;31(12):1211-3. doi: 10.2116/analsci.31.1211. Anal Sci. 2015. PMID: 26656807
-
Using time-lapse fluorescence microscopy to study gene regulation.Methods. 2019 Apr 15;159-160:138-145. doi: 10.1016/j.ymeth.2018.12.010. Epub 2018 Dec 29. Methods. 2019. PMID: 30599195 Free PMC article. Review.
-
Measuring transcription dynamics in living cells using a photobleaching approach.Methods. 2017 May 1;120:58-64. doi: 10.1016/j.ymeth.2017.04.007. Epub 2017 Apr 21. Methods. 2017. PMID: 28434903 Review.
Cited by
-
Increased COX-1 expression in benign prostate epithelial cells is triggered by mitochondrial dysfunction.Am J Clin Exp Urol. 2022 Aug 15;10(4):234-245. eCollection 2022. Am J Clin Exp Urol. 2022. PMID: 36051613 Free PMC article.
-
Poisson pre-processing of nonstationary photonic signals: Signals with equality between mean and variance.PLoS One. 2017 Dec 7;12(12):e0188622. doi: 10.1371/journal.pone.0188622. eCollection 2017. PLoS One. 2017. PMID: 29216207 Free PMC article.
-
Quantitative Imaging of Morphometric and Metabolic Signatures Reveals Heterogeneity in Drug Response of Three-Dimensional Mammary Tumor Spheroids.Mol Imaging Biol. 2019 Jun;21(3):436-446. doi: 10.1007/s11307-019-01324-7. Mol Imaging Biol. 2019. PMID: 30805885
-
Targeting metabolic plasticity in breast cancer cells via mitochondrial complex I modulation.Breast Cancer Res Treat. 2015 Feb;150(1):43-56. doi: 10.1007/s10549-015-3304-8. Epub 2015 Feb 13. Breast Cancer Res Treat. 2015. PMID: 25677747 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

