Lizards and LINEs: selection and demography affect the fate of L1 retrotransposons in the genome of the green anole (Anolis carolinensis)
- PMID: 24013105
- PMCID: PMC3787681
- DOI: 10.1093/gbe/evt133
Lizards and LINEs: selection and demography affect the fate of L1 retrotransposons in the genome of the green anole (Anolis carolinensis)
Erratum in
-
Lizards and LINEs: Selection and Demography Affect the Fate of L1 Retrotransposons in the Genome of the Green Anole (Anolis carolinensis).Genome Biol Evol. 2013 Jan;5(10):2019. doi: 10.1093/gbe/evt167. Genome Biol Evol. 2013. PMID: 27798071 Free PMC article. No abstract available.
Abstract
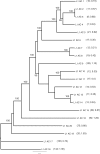
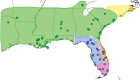
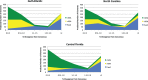
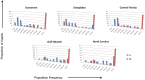
Autonomous retrotransposons lacking long terminal repeats (LTR) account for much of the variation in genome size and structure among vertebrates. Mammalian genomes contain hundreds of thousands of non-LTR retrotransposon copies, mostly resulting from the amplification of a single clade known as L1. The genomes of teleost fish and squamate reptiles contain a much more diverse array of non-LTR retrotransposon families, whereas copy number is relatively low. The majority of non-LTR retrotransposon insertions in nonmammalian vertebrates also appear to be very recent, suggesting strong purifying selection limits the accumulation of non-LTR retrotransposon copies. It is however unclear whether this turnover model, originally proposed in Drosophila, applies to nonmammalian vertebrates. Here, we studied the population dynamics of L1 in the green anole lizard (Anolis carolinensis). We found that although most L1 elements are recent in this genome, truncated insertions accumulate readily, and many are fixed at both the population and species level. In contrast, full-length L1 insertions are found at lower population frequencies, suggesting that the turnover model only applies to longer L1 elements in Anolis. We also found that full-length L1 inserts are more likely to be fixed in populations of small effective size, suggesting that the strength of purifying selection against deleterious alleles is highly dependent on host demographic history. Similar mechanisms seem to be controlling the fate of non-LTR retrotransposons in both Anolis and teleostean fish, which suggests that mammals have considerably diverged from the ancestral vertebrate in terms of how they interact with their intragenomic parasites.
Keywords: Anolis carolinensis; green anole; non-LTR retrotransposon; population genomics.
Figures

Similar articles
-
The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals.Mol Biol Evol. 2009 Aug;26(8):1811-22. doi: 10.1093/molbev/msp090. Epub 2009 May 6. Mol Biol Evol. 2009. PMID: 19420048
-
Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback.Genome Biol Evol. 2012;4(5):687-702. doi: 10.1093/gbe/evs044. Epub 2012 Apr 25. Genome Biol Evol. 2012. PMID: 22534163 Free PMC article.
-
The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis.Gene. 2009 Jul 15;441(1-2):111-8. doi: 10.1016/j.gene.2008.11.030. Epub 2008 Dec 14. Gene. 2009. PMID: 19118606
-
Co-evolution of plant LTR-retrotransposons and their host genomes.Protein Cell. 2013 Jul;4(7):493-501. doi: 10.1007/s13238-013-3037-6. Epub 2013 Jun 23. Protein Cell. 2013. PMID: 23794032 Free PMC article. Review.
-
LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model.Cytogenet Genome Res. 2005;110(1-4):91-107. doi: 10.1159/000084941. Cytogenet Genome Res. 2005. PMID: 16093661 Review.
Cited by
-
The genomic proliferation of transposable elements in colonizing populations: Schistosoma mansoni in the new world.Genetica. 2015 Jun;143(3):287-98. doi: 10.1007/s10709-015-9825-6. Epub 2015 Feb 14. Genetica. 2015. PMID: 25681233
-
The State of Squamate Genomics: Past, Present, and Future of Genome Research in the Most Speciose Terrestrial Vertebrate Order.Genes (Basel). 2023 Jul 1;14(7):1387. doi: 10.3390/genes14071387. Genes (Basel). 2023. PMID: 37510292 Free PMC article. Review.
-
Comparative Genomics Reveals Accelerated Evolution in Conserved Pathways during the Diversification of Anole Lizards.Genome Biol Evol. 2018 Feb 1;10(2):489-506. doi: 10.1093/gbe/evy013. Genome Biol Evol. 2018. PMID: 29360978 Free PMC article.
-
LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life.Genome Biol Evol. 2016 Dec 14;8(11):3301-3322. doi: 10.1093/gbe/evw243. Genome Biol Evol. 2016. PMID: 27702814 Free PMC article.
-
Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals.Nat Commun. 2018 Jul 17;9(1):2774. doi: 10.1038/s41467-018-05279-1. Nat Commun. 2018. PMID: 30018307 Free PMC article.
References
-
- Biemont C, et al. Population dynamics of the copia, mdg1, mdg3, gypsy, and P transposable elements in a natural population of Drosophila melanogaster. Genet Res. 1994;63:197–212. - PubMed
-
- Biemont C, et al. Maintenance of transposable element copy number in natural populations of Drosophila melanogaster and D. simulans. Genetica. 1997;100:161–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

