Noncoding RNA in oncogenesis: a new era of identifying key players
- PMID: 24013378
- PMCID: PMC3794782
- DOI: 10.3390/ijms140918319
Noncoding RNA in oncogenesis: a new era of identifying key players
Abstract
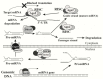
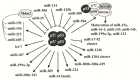
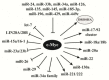
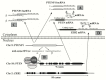
New discoveries and accelerating progresses in the field of noncoding RNAs (ncRNAs) continuously challenges our deep-rooted doctrines in biology and sometimes our imagination. A growing body of evidence indicates that ncRNAs are important players in oncogenesis. While a stunning list of ncRNAs has been discovered, only a small portion of them has been examined for their biological activities and very few have been characterized for the molecular mechanisms of their action. To date, ncRNAs have been shown to regulate a wide range of biological processes, including chromatin remodeling, gene transcription, mRNA translation and protein function. Dysregulation of ncRNAs contributes to the pathogenesis of a variety of cancers and aberrant ncRNA expression has a high potential to be prognostic in some cancers. Thus, a new cancer research era has begun to identify novel key players of ncRNAs in oncogenesis. In this review, we will first discuss the function and regulation of miRNAs, especially focusing on the interplay between miRNAs and several key cancer genes, including p53, PTEN and c-Myc. We will then summarize the research of long ncRNAs (lncRNAs) in cancers. In this part, we will discuss the lncRNAs in four categories based on their activities, including regulating gene expression, acting as miRNA decoys, mediating mRNA translation, and modulating protein activities. At the end, we will also discuss recently unraveled activities of circular RNAs (circRNAs).
Figures
References
-
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
-
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

