The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo
- PMID: 24013501
- PMCID: PMC3778241
- DOI: 10.1101/gad.224386.113
The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo
Abstract
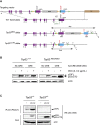
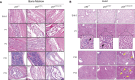
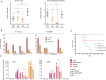
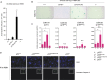
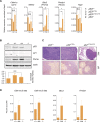
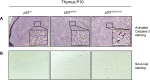
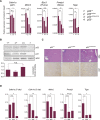
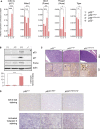
The p53 tumor suppressor is a transcription factor that mediates varied cellular responses. The C terminus of p53 is subjected to multiple and diverse post-translational modifications. An attractive hypothesis is that differing sets of combinatorial modifications therein determine distinct cellular outcomes. To address this in vivo, a Trp53(ΔCTD/ΔCTD) mouse was generated in which the endogenous p53 is targeted and replaced with a truncated mutant lacking the C-terminal 24 amino acids. These Trp53(ΔCTD/ΔCTD) mice die within 2 wk post-partum with hematopoietic failure and impaired cerebellar development. Intriguingly, the C terminus acts via three distinct mechanisms to control p53-dependent gene expression depending on the tissue. First, in the bone marrow and thymus, the C terminus dampens p53 activity. Increased senescence in the Trp53(ΔCTD/ΔCTD) bone marrow is accompanied by up-regulation of Cdkn1 (p21). In the thymus, the C-terminal domain negatively regulates p53-dependent gene expression by inhibiting promoter occupancy. Here, the hyperactive p53(ΔCTD) induces apoptosis via enhanced expression of the proapoptotic Bbc3 (Puma) and Pmaip1 (Noxa). In the liver, a second mechanism prevails, since p53(ΔCTD) has wild-type DNA binding but impaired gene expression. Thus, the C terminus of p53 is needed in liver cells at a step subsequent to DNA binding. Finally, in the spleen, the C terminus controls p53 protein levels, with the overexpressed p53(ΔCTD) showing hyperactivity for gene expression. Thus, the C terminus of p53 regulates gene expression via multiple mechanisms depending on the tissue and target, and this leads to specific phenotypic effects in vivo.
Keywords: C terminus; gene expression; hematopoiesis; mouse; p53; tissue specificity.
Figures

Similar articles
-
p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa.Cell Rep. 2013 May 30;3(5):1339-45. doi: 10.1016/j.celrep.2013.04.012. Epub 2013 May 9. Cell Rep. 2013. PMID: 23665218
-
The C terminus of p53 family proteins is a cell fate determinant.Mol Cell Biol. 2005 Mar;25(5):2014-30. doi: 10.1128/MCB.25.5.2014-2030.2005. Mol Cell Biol. 2005. PMID: 15713654 Free PMC article.
-
Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity.Mol Cell Biol. 2005 Jul;25(13):5389-95. doi: 10.1128/MCB.25.13.5389-5395.2005. Mol Cell Biol. 2005. PMID: 15964796 Free PMC article.
-
Signaling to p53: breaking the posttranslational modification code.Pathol Biol (Paris). 2000 Apr;48(3):227-45. Pathol Biol (Paris). 2000. PMID: 10858956 Review.
-
Post-translational modifications and activation of p53 by genotoxic stresses.Eur J Biochem. 2001 May;268(10):2764-72. doi: 10.1046/j.1432-1327.2001.02225.x. Eur J Biochem. 2001. PMID: 11358490 Review.
Cited by
-
The Tail That Wags the Dog: How the Disordered C-Terminal Domain Controls the Transcriptional Activities of the p53 Tumor-Suppressor Protein.Trends Biochem Sci. 2016 Dec;41(12):1022-1034. doi: 10.1016/j.tibs.2016.08.011. Epub 2016 Sep 23. Trends Biochem Sci. 2016. PMID: 27669647 Free PMC article. Review.
-
Navigating the complexity of p53-DNA binding: implications for cancer therapy.Biophys Rev. 2024 Jul 11;16(4):479-496. doi: 10.1007/s12551-024-01207-4. eCollection 2024 Aug. Biophys Rev. 2024. PMID: 39309126 Free PMC article. Review.
-
Intra molecular interactions in the regulation of p53 pathway.Transl Cancer Res. 2016 Dec;5(6):639-649. doi: 10.21037/tcr.2016.09.23. Transl Cancer Res. 2016. PMID: 30613472 Free PMC article.
-
Evidence for allosteric effects on p53 oligomerization induced by phosphorylation.Protein Sci. 2018 Feb;27(2):523-530. doi: 10.1002/pro.3344. Epub 2017 Dec 8. Protein Sci. 2018. PMID: 29124793 Free PMC article.
-
Mechanisms of transcriptional regulation by p53.Cell Death Differ. 2018 Jan;25(1):133-143. doi: 10.1038/cdd.2017.174. Epub 2017 Nov 10. Cell Death Differ. 2018. PMID: 29125602 Free PMC article. Review.
References
-
- Ahn J, Prives C 2001. The C-terminus of p53: The more you learn the less you know. Nat Struct Biol 8: 730–732 - PubMed
-
- Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR 1995. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol 5: 931–936 - PubMed
-
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
