Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries
- PMID: 24013502
- PMCID: PMC3778242
- DOI: 10.1101/gad.221010.113
Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries
Abstract
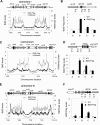
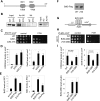
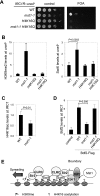
Heterochromatin spreading leads to the silencing of genes within its path, and boundary elements have evolved to constrain such spreading. In fission yeast, heterochromatin at centromeres I and III is flanked by inverted repeats termed IRCs, which are required for proper boundary functions. However, the mechanisms by which IRCs prevent heterochromatin spreading are unknown. Here, we identified Bdf2, which is homologous to the mammalian bromodomain and extraterminal (BET) family double bromodomain proteins involved in diverse types of cancers, as a factor required for proper boundary function at IRCs. Bdf2 is enriched at IRCs through its interaction with the boundary protein Epe1. The bromodomains of Bdf2 recognize acetylated histone H4 tails and antagonize Sir2-mediated deacetylation of histone H4K16. Furthermore, abolishing H4K16 acetylation (H4K16ac) with an H4K16R mutation promotes heterochromatin spreading, and mimicking H4K16ac by an H4K16Q mutation blocks heterochromatin spreading at IRCs. Our results thus illustrate a mechanism of establishing chromosome boundaries at specific sites through the recruitment of a factor that protects euchromatic histone modifications. They also reveal a previously unappreciated function of H4K16ac in cooperation with H3K9 methylation to regulate heterochromatin spreading.
Keywords: BET; H4K16; acetylation; boundary; heterochromatin.
Figures




Similar articles
-
Anti-silencing factor Epe1 associates with SAGA to regulate transcription within heterochromatin.Genes Dev. 2019 Jan 1;33(1-2):116-126. doi: 10.1101/gad.318030.118. Epub 2018 Dec 20. Genes Dev. 2019. PMID: 30573453 Free PMC article.
-
Intrinsic Toxicity of Unchecked Heterochromatin Spread Is Suppressed by Redundant Chromatin Boundary Functions in Schizosacchromyces pombe.G3 (Bethesda). 2015 May 8;5(7):1453-61. doi: 10.1534/g3.115.018663. G3 (Bethesda). 2015. PMID: 25957277 Free PMC article.
-
The histone chaperone FACT facilitates heterochromatin spreading by regulating histone turnover and H3K9 methylation states.Cell Rep. 2021 Nov 2;37(5):109944. doi: 10.1016/j.celrep.2021.109944. Cell Rep. 2021. PMID: 34731638 Free PMC article.
-
Destabilizing heterochromatin: Does Swi6/HP1 make the choice?Mol Cell. 2006 Jun 23;22(6):709-710. doi: 10.1016/j.molcel.2006.06.004. Mol Cell. 2006. PMID: 16793539 Review.
-
Unprogrammed epigenetic variation mediated by stochastic formation of ectopic heterochromatin.Curr Genet. 2020 Apr;66(2):319-325. doi: 10.1007/s00294-019-01031-4. Epub 2019 Oct 9. Curr Genet. 2020. PMID: 31598751 Review.
Cited by
-
The Fission Yeast Mating-Type Switching Motto: "One-for-Two" and "Two-for-One".Microbiol Mol Biol Rev. 2023 Mar 21;87(1):e0000821. doi: 10.1128/mmbr.00008-21. Epub 2023 Jan 11. Microbiol Mol Biol Rev. 2023. PMID: 36629411 Free PMC article. Review.
-
TOR complex 2 in fission yeast is required for chromatin-mediated gene silencing and assembly of heterochromatic domains at subtelomeres.J Biol Chem. 2018 May 25;293(21):8138-8150. doi: 10.1074/jbc.RA118.002270. Epub 2018 Apr 9. J Biol Chem. 2018. PMID: 29632066 Free PMC article.
-
A meiotic driver hijacks an epigenetic reader to disrupt mitosis in noncarrier offspring.Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2408347121. doi: 10.1073/pnas.2408347121. Epub 2024 Nov 1. Proc Natl Acad Sci U S A. 2024. PMID: 39485795 Free PMC article.
-
The cAMP signaling pathway regulates Epe1 protein levels and heterochromatin assembly.PLoS Genet. 2022 Feb 16;18(2):e1010049. doi: 10.1371/journal.pgen.1010049. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35171902 Free PMC article.
-
Chromosome boundary elements and regulation of heterochromatin spreading.Cell Mol Life Sci. 2014 Dec;71(24):4841-52. doi: 10.1007/s00018-014-1725-x. Epub 2014 Sep 7. Cell Mol Life Sci. 2014. PMID: 25192661 Free PMC article. Review.
References
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
-
- Beisel C, Paro R 2011. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet 12: 123–135 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
