Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection
- PMID: 24013561
- PMCID: PMC3905848
- DOI: 10.1093/nar/gkt802
Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection
Abstract
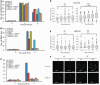
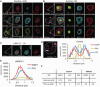
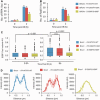
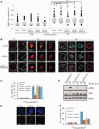
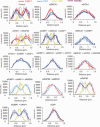
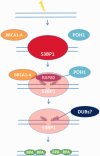
In G2 phase cells, DNA double-strand break repair switches from DNA non-homologous end-joining to homologous recombination. This switch demands the promotion of resection. We examine the changes in 53BP1 and RAP80 ionizing radiation induced foci (IRIF) in G2 phase, as these are factors that restrict resection. We observed a 2-fold increase in the volume of 53BP1 foci by 8 h, which is not seen in G1 cells. Additionally, an IRIF core devoid of 53BP1 arises where RPA foci form, with BRCA1 IRIF forming between 53BP1 and replication protein A (RPA). Ubiquitin chains assessed using α-FK2 antibodies are similarly repositioned. Repositioning of all these components requires BRCA1's BRCT but not the ring finger domain. 53BP1, RAP80 and ubiquitin chains are enlarged following POH1 depletion by small interfering RNA, but a devoid core does not form and RPA foci formation is impaired. Co-depletion of POH1 and RAP80, BRCC36 or ABRAXAS allows establishment of the 53BP1 and ubiquitin chain-devoid core. Thus, the barriers posed by 53BP1 and RAP80 are relieved by BRCA1 and POH1, respectively. Analysis of combined depletions shows that these represent distinct but interfacing barriers to promote loss of ubiquitin chains in the IRIF core, which is required for subsequent resection. We propose a model whereby BRCA1 impacts on 53BP1 to allow access of POH1 to RAP80. POH1-dependent removal of RAP80 within the IRIF core enables degradation of ubiquitin chains, which promotes loss of 53BP1. Thus, POH1 represents a novel component regulating the switch from non-homologous end-joining to homologous recombination.
Figures
Similar articles
-
DNA DSB repair pathway choice: an orchestrated handover mechanism.Br J Radiol. 2014 Mar;87(1035):20130685. doi: 10.1259/bjr.20130685. Br J Radiol. 2014. PMID: 24363387 Free PMC article. Review.
-
The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response.Cancer Res. 2007 Jul 15;67(14):6647-56. doi: 10.1158/0008-5472.CAN-07-0924. Epub 2007 Jul 9. Cancer Res. 2007. PMID: 17621610 Free PMC article.
-
A small ubiquitin binding domain inhibits ubiquitin-dependent protein recruitment to DNA repair foci.Cell Cycle. 2013 Dec 15;12(24):3749-58. doi: 10.4161/cc.26640. Epub 2013 Oct 3. Cell Cycle. 2013. PMID: 24107634 Free PMC article.
-
Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage.Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20759-63. doi: 10.1073/pnas.0710061104. Epub 2007 Dec 5. Proc Natl Acad Sci U S A. 2007. PMID: 18077395 Free PMC article.
-
RAP80, ubiquitin and SUMO in the DNA damage response.J Mol Med (Berl). 2017 Aug;95(8):799-807. doi: 10.1007/s00109-017-1561-1. Epub 2017 Jul 5. J Mol Med (Berl). 2017. PMID: 28681078 Free PMC article. Review.
Cited by
-
Regulation of Histone Ubiquitination in Response to DNA Double Strand Breaks.Cells. 2020 Jul 16;9(7):1699. doi: 10.3390/cells9071699. Cells. 2020. PMID: 32708614 Free PMC article. Review.
-
DNA DSB repair pathway choice: an orchestrated handover mechanism.Br J Radiol. 2014 Mar;87(1035):20130685. doi: 10.1259/bjr.20130685. Br J Radiol. 2014. PMID: 24363387 Free PMC article. Review.
-
Role of Deubiquitinating Enzymes in DNA Repair.Mol Cell Biol. 2015 Dec 7;36(4):524-44. doi: 10.1128/MCB.00847-15. Print 2016 Feb 15. Mol Cell Biol. 2015. PMID: 26644404 Free PMC article. Review.
-
BRCA1-A and BRISC: Multifunctional Molecular Machines for Ubiquitin Signaling.Biomolecules. 2020 Oct 31;10(11):1503. doi: 10.3390/biom10111503. Biomolecules. 2020. PMID: 33142801 Free PMC article. Review.
-
Writers, Readers, and Erasers of Histone Ubiquitylation in DNA Double-Strand Break Repair.Front Genet. 2016 Jun 28;7:122. doi: 10.3389/fgene.2016.00122. eCollection 2016. Front Genet. 2016. PMID: 27446204 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

