Suppression of HPV-16 late L1 5'-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs
- PMID: 24013563
- PMCID: PMC3905901
- DOI: 10.1093/nar/gkt803
Suppression of HPV-16 late L1 5'-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs
Abstract
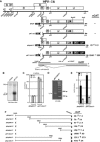
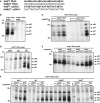
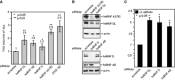
Human papillomavirus type 16 (HPV-16) 5'-splice site SD3632 is used exclusively to produce late L1 mRNAs. We identified a 34-nt splicing inhibitory element located immediately upstream of HPV-16 late 5'-splice site SD3632. Two AUAGUA motifs located in these 34 nt inhibited SD3632. Two nucleotide substitutions in each of the HPV-16 specific AUAGUA motifs alleviated splicing inhibition and induced late L1 mRNA production from episomal forms of the HPV-16 genome in primary human keratinocytes. The AUAGUA motifs bind specifically not only to the heterogeneous nuclear RNP (hnRNP) D family of RNA-binding proteins including hnRNP D/AUF, hnRNP DL and hnRNP AB but also to hnRNP A2/B1. Knock-down of these proteins induced HPV-16 late L1 mRNA expression, and overexpression of hnRNP A2/B1, hnRNP AB, hnRNP DL and the two hnRNP D isoforms hnRNP D37 and hnRNP D40 further suppressed L1 mRNA expression. This inhibition may allow HPV-16 to hide from the immune system and establish long-term persistent infections with enhanced risk at progressing to cancer. There is an inverse correlation between expression of hnRNP D proteins and hnRNP A2/B1 and HPV-16 L1 production in the cervical epithelium, as well as in cervical cancer, supporting the conclusion that hnRNP D proteins and A2/B1 inhibit HPV-16 L1 mRNA production.
Figures






References
-
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. - PubMed
-
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. - PubMed
-
- Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur. J. Cancer. 2001;37(Suppl. 8):24–66. - PubMed
-
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, et al. A review of human carcinogens—part B: biological agents. Lancet Oncol. 2009;10:321–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

