Identification and characterization of non-cellulose-producing mutants of Gluconacetobacter hansenii generated by Tn5 transposon mutagenesis
- PMID: 24013627
- PMCID: PMC3811599
- DOI: 10.1128/JB.00767-13
Identification and characterization of non-cellulose-producing mutants of Gluconacetobacter hansenii generated by Tn5 transposon mutagenesis
Abstract
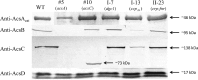
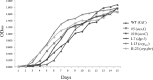
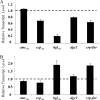
The acs operon of Gluconacetobacter is thought to encode AcsA, AcsB, AcsC, and AcsD proteins that constitute the cellulose synthase complex, required for the synthesis and secretion of crystalline cellulose microfibrils. A few other genes have been shown to be involved in this process, but their precise role is unclear. We report here the use of Tn5 transposon insertion mutagenesis to identify and characterize six non-cellulose-producing (Cel(-)) mutants of Gluconacetobacter hansenii ATCC 23769. The genes disrupted were acsA, acsC, ccpAx (encoding cellulose-complementing protein [the subscript "Ax" indicates genes from organisms formerly classified as Acetobacter xylinum]), dgc1 (encoding guanylate dicyclase), and crp-fnr (encoding a cyclic AMP receptor protein/fumarate nitrate reductase transcriptional regulator). Protein blot analysis revealed that (i) AcsB and AcsC were absent in the acsA mutant, (ii) the levels of AcsB and AcsC were significantly reduced in the ccpAx mutant, and (iii) the level of AcsD was not affected in any of the Cel(-) mutants. Promoter analysis showed that the acs operon does not include acsD, unlike the organization of the acs operon of several strains of closely related Gluconacetobacter xylinus. Complementation experiments confirmed that the gene disrupted in each Cel(-) mutant was responsible for the phenotype. Quantitative real-time PCR and protein blotting results suggest that the transcription of bglAx (encoding β-glucosidase and located immediately downstream from acsD) was strongly dependent on Crp/Fnr. A bglAx knockout mutant, generated via homologous recombination, produced only ∼16% of the wild-type cellulose level. Since the crp-fnr mutant did not produce any cellulose, Crp/Fnr may regulate the expression of other gene(s) involved in cellulose biosynthesis.
Figures



References
-
- Römling U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205–212 - PubMed
-
- Valla S, Ertesvåg H, Tonouchi N, Fjærvik E. 2009. Bacterial cellulose production: biosynthesis and applications, p 43–77 In Rehm BHA. (ed), Microbial production of biopolymers and polymer precursors: applications and perspectives. Caister Academic Press, Norfolk, United Kingdom
-
- Kawano S, Tajima K, Uemori Y, Yamashta H, Erata T, Munekata M, Takai M. 2002. Cloning of cellulose synthesis related gene from Acetobacter xylinum ATCC 23769 and ATCC 53582: comparison of cellulose synthetic ability between strains. DNA Res. 9:149–156 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

