The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination
- PMID: 24013629
- PMCID: PMC3811594
- DOI: 10.1128/JB.00901-13
The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination
Abstract
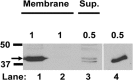
The Gram-positive, anaerobic, spore-forming bacterium Clostridium perfringens causes a variety of diseases in both humans and animals, and spore germination is thought to be the first stage of C. perfringens infection. Previous studies have indicated that the germinant receptor (GR) proteins encoded by the bicistronic gerKA-gerKC operon as well as the proteins encoded by the gerKB and gerAA genes are required for normal germination of C. perfringens spores. We now report the individual role of these GR proteins by analyzing the germination of strains carrying mutations in gerKA, gerKC, or both gerKB and gerAA. Western blot analysis was also used to determine the location and numbers of GerKC proteins in spores. Conclusions from this work include the following: (i) gerKC mutant spores germinate extremely poorly with KCl, l-asparagine, a mixture of asparagine and KCl, or NaPi; (ii) gerKC spores germinate significantly more slowly than wild-type and other GR mutant spores with a 1:1 chelate of Ca(2+) and dipicolinic acid and very slightly more slowly with dodecylamine; (iii) the germination defects in gerKC spores are largely restored by expressing the wild-type gerKA-gerKC operon in trans; (iv) GerKC is required for the spores' viability, almost certainly because of the gerKC spores' poor germination; and (v) GerKC is located in the spores' inner membrane, with ∼250 molecules/spore. Collectively, these results indicate that GerKC is the main GR protein required for nutrient and nonnutrient germination of spores of C. perfringens food-poisoning isolates.
Figures






Similar articles
-
Characterization of germinants and their receptors for spores of non-food-borne Clostridium perfringens strain F4969.Microbiology (Reading). 2016 Nov;162(11):1972-1983. doi: 10.1099/mic.0.000378. Epub 2016 Sep 29. Microbiology (Reading). 2016. PMID: 27692042
-
New amino acid germinants for spores of the enterotoxigenic Clostridium perfringens type A isolates.Food Microbiol. 2014 Dec;44:24-33. doi: 10.1016/j.fm.2014.04.011. Epub 2014 May 6. Food Microbiol. 2014. PMID: 25084641
-
l-lysine (pH 6.0) induces germination of spores of Clostridium perfringens type F isolates carrying chromosomal or plasmid-borne enterotoxin gene.Microb Pathog. 2018 Oct;123:227-232. doi: 10.1016/j.micpath.2018.07.022. Epub 2018 Jul 18. Microb Pathog. 2018. PMID: 30031038
-
Germination of spores of Bacillus species: what we know and do not know.J Bacteriol. 2014 Apr;196(7):1297-305. doi: 10.1128/JB.01455-13. Epub 2014 Jan 31. J Bacteriol. 2014. PMID: 24488313 Free PMC article. Review.
-
Updates to Clostridium difficile Spore Germination.J Bacteriol. 2018 Jul 25;200(16):e00218-18. doi: 10.1128/JB.00218-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29760211 Free PMC article. Review.
Cited by
-
Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia.Antibiotics (Basel). 2022 Aug 29;11(9):1165. doi: 10.3390/antibiotics11091165. Antibiotics (Basel). 2022. PMID: 36139945 Free PMC article. Review.
-
The orphan germinant receptor protein GerXAO (but not GerX3b) is essential for L-alanine induced germination in Clostridium botulinum Group II.Sci Rep. 2018 May 4;8(1):7060. doi: 10.1038/s41598-018-25411-x. Sci Rep. 2018. PMID: 29728678 Free PMC article.
-
Impact of pH and High-Pressure Pasteurization on the Germination and Development of Clostridium perfringens Spores under Hyperbaric Storage versus Refrigeration.Foods. 2024 Jun 11;13(12):1832. doi: 10.3390/foods13121832. Foods. 2024. PMID: 38928774 Free PMC article.
-
Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis.Mob DNA. 2014 Jan 13;5(1):2. doi: 10.1186/1759-8753-5-2. Mob DNA. 2014. PMID: 24410776 Free PMC article.
-
Identification of Germinants and Expression of Germination Genes in Clostridium perfringens Strains Isolated from Diarrheic Animals.Pathogens. 2024 Feb 22;13(3):194. doi: 10.3390/pathogens13030194. Pathogens. 2024. PMID: 38535537 Free PMC article.
References
-
- McClane BA. 2007. Clostridium perfringens, p 423–444 In Doyle MP, Beuchat LR. (ed), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC
-
- McDonnell JL. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p 477–517 In Dorner F, Drews J. (ed), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom
-
- McClane B, Uzal FA, Miyakawa MF, Lyerly D, Wilkins T. 2006. The enterotoxic clostridia, p 698–752 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

