Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence
- PMID: 24013631
- PMCID: PMC3811582
- DOI: 10.1128/JB.00769-13
Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence
Abstract
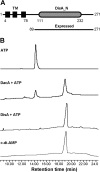
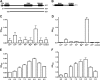
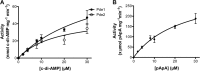
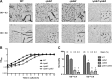
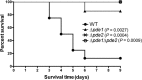
Cyclic di-AMP (c-di-AMP) and cyclic di-GMP (c-di-GMP) are signaling molecules that play important roles in bacterial biology and pathogenesis. However, these nucleotides have not been explored in Streptococcus pneumoniae, an important bacterial pathogen. In this study, we characterized the c-di-AMP-associated genes of S. pneumoniae. The results showed that SPD_1392 (DacA) is a diadenylate cyclase that converts ATP to c-di-AMP. Both SPD_2032 (Pde1) and SPD_1153 (Pde2), which belong to the DHH subfamily 1 proteins, displayed c-di-AMP phosphodiesterase activity. Pde1 cleaved c-di-AMP into phosphoadenylyl adenosine (pApA), whereas Pde2 directly hydrolyzed c-di-AMP into AMP. Additionally, Pde2, but not Pde1, degraded pApA into AMP. Our results also demonstrated that both Pde1 and Pde2 played roles in bacterial growth, resistance to UV treatment, and virulence in a mouse pneumonia model. These results indicate that c-di-AMP homeostasis is essential for pneumococcal biology and disease.
Figures


References
-
- Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

