Finely tuned regulation of the aromatic amine degradation pathway in Escherichia coli
- PMID: 24013633
- PMCID: PMC3811602
- DOI: 10.1128/JB.00837-13
Finely tuned regulation of the aromatic amine degradation pathway in Escherichia coli
Abstract
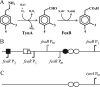
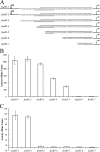
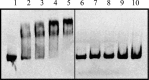
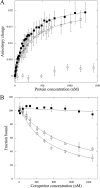
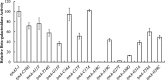
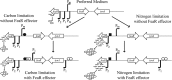
FeaR is an AraC family regulator that activates transcription of the tynA and feaB genes in Escherichia coli. TynA is a periplasmic topaquinone- and copper-containing amine oxidase, and FeaB is a cytosolic NAD-linked aldehyde dehydrogenase. Phenylethylamine, tyramine, and dopamine are oxidized by TynA to the corresponding aldehydes, releasing one equivalent of H2O2 and NH3. The aldehydes can be oxidized to carboxylic acids by FeaB, and (in the case of phenylacetate) can be further degraded to enter central metabolism. Thus, phenylethylamine can be used as a carbon and nitrogen source, while tyramine and dopamine can be used only as sources of nitrogen. Using genetic, biochemical and computational approaches, we show that the FeaR binding site is a TGNCA-N8-AAA motif that occurs in 2 copies in the tynA and feaB promoters. We show that the coactivator for FeaR is the product rather than the substrate of the TynA reaction. The feaR gene is upregulated by carbon or nitrogen limitation, which we propose reflects regulation of feaR by the cyclic AMP receptor protein (CRP) and the nitrogen assimilation control protein (NAC), respectively. In carbon-limited cells grown in the presence of a TynA substrate, tynA and feaB are induced, whereas in nitrogen-limited cells, only the tynA promoter is induced. We propose that tynA and feaB expression is finely tuned to provide the FeaB activity that is required for carbon source utilization and the TynA activity required for nitrogen and carbon source utilization.
Figures



References
-
- Klinman JP. 2003. The multi-functional topa-quinone copper amine oxidases. Biochim. Biophys. Acta 1647:131–137 - PubMed
-
- Ferrandez A, Prieto MA, Garcia JL, Diaz E. 1997. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 406:23–27 - PubMed
-
- Hanlon SP, Hill TK, Flavell MA, Stringfellow JM, Cooper RA. 1997. 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. J. Gen. Microbiol. 143:513–518 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

