Circadian clock regulation of the cell cycle in the zebrafish intestine
- PMID: 24013905
- PMCID: PMC3754960
- DOI: 10.1371/journal.pone.0073209
Circadian clock regulation of the cell cycle in the zebrafish intestine
Abstract
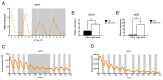
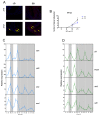
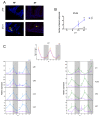
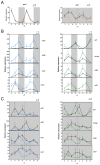
The circadian clock controls cell proliferation in a number of healthy tissues where cell renewal and regeneration are critical for normal physiological function. The intestine is an organ that typically undergoes regular cycles of cell division, differentiation and apoptosis as part of its role in digestion and nutrient absorption. The aim of this study was to explore circadian clock regulation of cell proliferation and cell cycle gene expression in the zebrafish intestine. Here we show that the zebrafish gut contains a directly light-entrainable circadian pacemaker, which regulates the daily timing of mitosis. Furthermore, this intestinal clock controls the expression of key cell cycle regulators, such as cdc2, wee1, p21, PCNA and cdk2, but only weakly influences cyclin B1, cyclin B2 and cyclin E1 expression. Interestingly, food deprivation has little impact on circadian clock function in the gut, but dramatically reduces cell proliferation, as well as cell cycle gene expression in this tissue. Timed feeding under constant dark conditions is able to drive rhythmic expression not only of circadian clock genes, but also of several cell cycle genes, suggesting that food can entrain the clock, as well as the cell cycle in the intestine. Rather surprisingly, we found that timed feeding is critical for high amplitude rhythms in cell cycle gene expression, even when zebrafish are maintained on a light-dark cycle. Together these results suggest that the intestinal clock integrates multiple rhythmic cues, including light and food, to function optimally.
Conflict of interest statement
Figures

References
-
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935-941. doi:10.1038/nature00965. PubMed: 12198538. - DOI - PubMed
-
- Panda S, Hogenesch JB, Kay SA (2002) Circadian rhythms from flies to human. Nature 417: 329-335. doi:10.1038/417329a. PubMed: 12015613. - DOI - PubMed
-
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE et al. (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544-556. doi:10.1038/nrg1633. PubMed: 15951747. - DOI - PMC - PubMed
-
- Hoogerwerf WA (2006) Biologic clocks and the gut. Curr Gastroenterol Rep 8: 353-359. doi:10.1007/s11894-006-0019-3. PubMed: 16968601. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

