The outer membrane TolC-like channel HgdD is part of tripartite resistance-nodulation-cell division (RND) efflux systems conferring multiple-drug resistance in the Cyanobacterium Anabaena sp. PCC7120
- PMID: 24014018
- PMCID: PMC3829430
- DOI: 10.1074/jbc.M113.495598
The outer membrane TolC-like channel HgdD is part of tripartite resistance-nodulation-cell division (RND) efflux systems conferring multiple-drug resistance in the Cyanobacterium Anabaena sp. PCC7120
Abstract
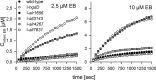
The TolC-like protein HgdD of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. PCC 7120 is part of multiple three-component "AB-D" systems spanning the inner and outer membranes and is involved in secretion of various compounds, including lipids, metabolites, antibiotics, and proteins. Several components of HgdD-dependent tripartite transport systems have been identified, but the diversity of inner membrane energizing systems is still unknown. Here we identified six putative resistance-nodulation-cell division (RND) type factors. Four of them are expressed during late exponential and stationary growth phase under normal growth conditions, whereas the other two are induced upon incubation with erythromycin or ethidium bromide. The constitutively expressed RND component Alr4267 has an atypical predicted topology, and a mutant strain (I-alr4267) shows a reduction in the content of monogalactosyldiacylglycerol as well as an altered filament shape. An insertion mutant of the ethidium bromide-induced all7631 did not show any significant phenotypic alteration under the conditions tested. Mutants of the constitutively expressed all3143 and alr1656 exhibited a Fox(-) phenotype. The phenotype of the insertion mutant I-all3143 parallels that of the I-hgdD mutant with respect to antibiotic sensitivity, lipid profile, and ethidium efflux. In addition, expression of the RND genes all3143 and all3144 partially complements the capability of Escherichia coli ΔacrAB to transport ethidium. We postulate that the RND transporter All3143 and the predicted membrane fusion protein All3144, as homologs of E. coli AcrB and AcrA, respectively, are major players for antibiotic resistance in Anabaena sp. PCC 7120.
Keywords: Antibiotic Resistance; Cyanobacteria; Membrane; Metabolite Transport; Multidrug Transporters; Nitrosative Stress; RND; TolC.
Figures







Similar articles
-
The TolC-like protein HgdD of the cyanobacterium Anabaena sp. PCC 7120 is involved in secondary metabolite export and antibiotic resistance.J Biol Chem. 2012 Nov 30;287(49):41126-38. doi: 10.1074/jbc.M112.396010. Epub 2012 Oct 15. J Biol Chem. 2012. PMID: 23071120 Free PMC article.
-
Secretome analysis of Anabaena sp. PCC 7120 and the involvement of the TolC-homologue HgdD in protein secretion.Environ Microbiol. 2015 Mar;17(3):767-80. doi: 10.1111/1462-2920.12516. Epub 2014 Jul 15. Environ Microbiol. 2015. PMID: 24890022
-
Structure-function analysis of the ATP-driven glycolipid efflux pump DevBCA reveals complex organization with TolC/HgdD.FEBS Lett. 2014 Jan 31;588(3):395-400. doi: 10.1016/j.febslet.2013.12.004. Epub 2013 Dec 17. FEBS Lett. 2014. PMID: 24361095
-
The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance.Curr Drug Targets. 2008 Sep;9(9):729-49. doi: 10.2174/138945008785747789. Curr Drug Targets. 2008. PMID: 18781920 Review.
-
AcrB: a mean, keen, drug efflux machine.Ann N Y Acad Sci. 2020 Jan;1459(1):38-68. doi: 10.1111/nyas.14239. Epub 2019 Oct 6. Ann N Y Acad Sci. 2020. PMID: 31588569 Review.
Cited by
-
Metabolic Responses, Uptake, and Export of Copper in Cyanobacteria.Biology (Basel). 2025 Jul 1;14(7):798. doi: 10.3390/biology14070798. Biology (Basel). 2025. PMID: 40723357 Free PMC article. Review.
-
Extracellular Vesicles: An Overlooked Secretion System in Cyanobacteria.Life (Basel). 2020 Jul 31;10(8):129. doi: 10.3390/life10080129. Life (Basel). 2020. PMID: 32751844 Free PMC article. Review.
-
Diversity and Evolution of Iron Uptake Pathways in Marine Cyanobacteria from the Perspective of the Coastal Strain Synechococcus sp. Strain PCC 7002.Appl Environ Microbiol. 2023 Jan 31;89(1):e0173222. doi: 10.1128/aem.01732-22. Epub 2022 Dec 20. Appl Environ Microbiol. 2023. PMID: 36533965 Free PMC article.
-
The ABC Transporter Components HgdB and HgdC are Important for Glycolipid Layer Composition and Function of Heterocysts in Anabaena sp. PCC 7120.Life (Basel). 2018 Jul 2;8(3):26. doi: 10.3390/life8030026. Life (Basel). 2018. PMID: 30004454 Free PMC article.
-
Structure-function of cyanobacterial outer-membrane protein, Slr1270: homolog of Escherichia coli drug export/colicin import protein, TolC.FEBS Lett. 2014 Nov 3;588(21):3793-801. doi: 10.1016/j.febslet.2014.08.028. Epub 2014 Sep 13. FEBS Lett. 2014. PMID: 25218435 Free PMC article.
References
-
- Piddock L. J. (2006) Multidrug-resistance efflux pumps. Not just for resistance. Nat. Rev. Microbiol. 4, 629–636 - PubMed
-
- Pos K. M. (2009) Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793 - PubMed
-
- Mirus O., Hahn A., Schleiff E. (2010) Outer Membrane Proteins. in Prokaryotic Cell Wall Compounds: Structure and Biochemistry (König H., Claus H., Varma A., eds) pp. 175–230, Springer-Verlag, Berlin
-
- Delepelaire P. (2004) Type I secretion in Gram-negative bacteria. Biochim. Biophys. Acta. 1694, 149–161 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

