Characterization of the AtsR hybrid sensor kinase phosphorelay pathway and identification of its response regulator in Burkholderia cenocepacia
- PMID: 24014026
- PMCID: PMC3798511
- DOI: 10.1074/jbc.M113.489914
Characterization of the AtsR hybrid sensor kinase phosphorelay pathway and identification of its response regulator in Burkholderia cenocepacia
Abstract
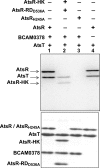
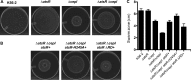
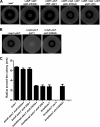
AtsR is a membrane-bound hybrid sensor kinase of Burkholderia cenocepacia that negatively regulates quorum sensing and virulence factors such as biofilm production, type 6-secretion, and protease secretion. Here we elucidate the mechanism of AtsR phosphorelay by site-directed mutagenesis of predicted histidine and aspartic acid phosphoacceptor residues. We demonstrate by in vitro phosphorylation that histidine 245 and aspartic acid 536 are conserved sites of phosphorylation in AtsR, and we also identify the cytosolic response regulator AtsT (BCAM0381) as a key component of the AtsR phosphorelay pathway. Monitoring the function of AtsR and its derivatives in vivo by measuring extracellular protease activity and swarming motility confirmed the in vitro phosphorylation results. Together we find that the AtsR receiver domain plays a fine-tuning role in determining the levels of phosphotransfer from its sensor kinase domain to the AtsT response regulator.
Keywords: Bacterial Protein Kinases; Cystic Fibrosis; Histidine Kinases; Macrophages; Phosphorylation; Protease; Transcription Regulation; Virulence Factors.
Figures






References
-
- Krell T., Lacal J., Busch A., Silva-Jiménez H., Guazzaroni M. E., Ramos J. L. (2010) Bacterial sensor kinases. Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 64, 539–559 - PubMed
-
- Mahenthiralingam E., Baldwin A., Dowson C. G. (2008) Burkholderia cepacia complex bacteria. Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

