Assessing the conformational changes of pb5, the receptor-binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA
- PMID: 24014030
- PMCID: PMC3798546
- DOI: 10.1074/jbc.M113.501536
Assessing the conformational changes of pb5, the receptor-binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA
Abstract
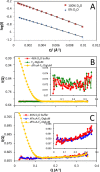
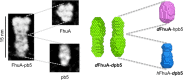
Within tailed bacteriophages, interaction of the receptor-binding protein (RBP) with the target cell triggers viral DNA ejection into the host cytoplasm. In the case of phage T5, the RBP pb5 and the receptor FhuA, an outer membrane protein of Escherichia coli, have been identified. Here, we use small angle neutron scattering and electron microscopy to investigate the FhuA-pb5 complex. Specific deuteration of one of the partners allows the complete masking in small angle neutron scattering of the surfactant and unlabeled proteins when the complex is solubilized in the fluorinated surfactant F6-DigluM. Thus, individual structures within a membrane protein complex can be described. The solution structure of FhuA agrees with its crystal structure; that of pb5 shows an elongated shape. Neither displays significant conformational changes upon interaction. The mechanism of signal transduction within phage T5 thus appears different from that of phages binding cell wall saccharides, for which structural information is available.
Keywords: Bacteriophage; Biophysics; Electron Microscopy (EM); Fluorinated Surfactant; Membrane Proteins; Small Angle Neutron Scattering; Viral Protein.
Figures





Similar articles
-
New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA.Biochimie. 2012 Sep;94(9):1982-9. doi: 10.1016/j.biochi.2012.05.021. Epub 2012 May 29. Biochimie. 2012. PMID: 22659573
-
Characterization of a high-affinity complex between the bacterial outer membrane protein FhuA and the phage T5 protein pb5.J Mol Biol. 2002 Apr 26;318(2):557-69. doi: 10.1016/S0022-2836(02)00089-X. J Mol Biol. 2002. PMID: 12051859
-
Structural basis for host recognition and superinfection exclusion by bacteriophage T5.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2211672119. doi: 10.1073/pnas.2211672119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215462 Free PMC article.
-
FhuA, an Escherichia coli outer membrane protein with a dual function of transporter and channel which mediates the transport of phage DNA.Biochimie. 1998 May-Jun;80(5-6):363-9. doi: 10.1016/s0300-9084(00)80004-8. Biochimie. 1998. PMID: 9782377 Review.
-
Energy-coupled transport through the outer membrane of Escherichia coli small deletions in the gating loop convert the FhuA transport protein into a diffusion channel.FEBS Lett. 1994 Jun 6;346(1):59-64. doi: 10.1016/0014-5793(94)00431-5. FEBS Lett. 1994. PMID: 7515827 Review.
Cited by
-
Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components.J Virol. 2014 Jan;88(2):1162-74. doi: 10.1128/JVI.02262-13. Epub 2013 Nov 6. J Virol. 2014. PMID: 24198424 Free PMC article.
-
Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages.J Virol. 2014 Jan;88(2):820-8. doi: 10.1128/JVI.02135-13. Epub 2013 Oct 23. J Virol. 2014. PMID: 24155371 Free PMC article.
-
Branched Lateral Tail Fiber Organization in T5-Like Bacteriophages DT57C and DT571/2 is Revealed by Genetic and Functional Analysis.Viruses. 2016 Jan 21;8(1):26. doi: 10.3390/v8010026. Viruses. 2016. PMID: 26805872 Free PMC article.
-
Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors.Viruses. 2020 May 6;12(5):512. doi: 10.3390/v12050512. Viruses. 2020. PMID: 32384698 Free PMC article. Review.
-
Development of a biosensor protein bullet as a fluorescent method for fast detection of Escherichia coli in drinking water.PLoS One. 2018 Jan 5;13(1):e0184277. doi: 10.1371/journal.pone.0184277. eCollection 2018. PLoS One. 2018. PMID: 29304041 Free PMC article.
References
-
- Soto C. M., Ratna B. R. (2010) Virus hybrids as nanomaterials for biotechnology. Curr. Opin. Biotechnol. 21, 426–438 - PubMed
-
- Maura D., Debarbieux L. (2011) Bacteriophages as twenty-first century antibacterial tools for food and medicine. Appl. Microbiol. Biotechnol. 90, 851–859 - PubMed
-
- Stone R. (2002) Bacteriophage therapy. Food and agriculture. Testing grounds for phage therapy. Science 298, 730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

