Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development
- PMID: 24014454
- PMCID: PMC3767924
- DOI: 10.1002/wdev.109
Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development
Abstract
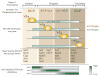
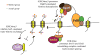
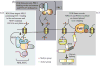
Set2 is a RNA polymerase II (RNAPII) associated histone methyltransferase involved in the cotranscriptional methylation of the H3 K36 residue (H3K36me). It is responsible for multiple degrees of methylation (mono-, di-, and trimethylation), each of which has a distinct functional consequence. The extent of methylation and its genomic distribution is determined by different factors that coordinate to achieve a functional outcome. In yeast, the Set2-mediated H3K36me is involved in suppressing histone exchange, preventing hyperacetylation and promoting maintenance of well-spaced chromatin structure over the coding regions. In metazoans, separation of this enzymatic activity affords greater functional diversity extending beyond the control of transcription elongation to developmental gene regulation. This review focuses on the molecular aspects of the Set2 distribution and function, and discusses the role played by H3 K36 methyl mark in organismal development.
Copyright © 2013 Wiley Periodicals, Inc.
Figures


References
-
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature genetics. 2004;36:900–905. - PubMed
-
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & development. 2004;18:2437–2468. - PubMed
-
- Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Current opinion in cell biology. 2004;16:263–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

