Initial neurogenesis in Drosophila
- PMID: 24014455
- PMCID: PMC3928071
- DOI: 10.1002/wdev.111
Initial neurogenesis in Drosophila
Erratum in
- Wiley Interdiscip Rev Dev Biol. 2013 Nov-Dec;2(6):823
-
Initial neurogenesis in Drosophila.Wiley Interdiscip Rev Dev Biol. 2013 Nov;2(6):823. doi: 10.1002/wdev.117. Epub 2013 Mar 22. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 29393596 No abstract available.
Abstract
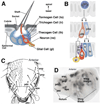
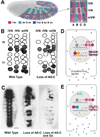
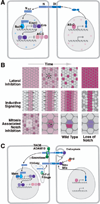
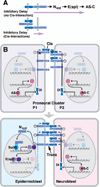
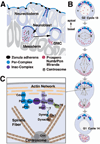
Early neurogenesis comprises the phase of nervous system development during which neural progenitor cells are born. In early development, the embryonic ectoderm is subdivided by a conserved signaling mechanism into two main domains, the epidermal ectoderm and the neurectoderm. Subsequently, cells of the neurectoderm are internalized and form a cell layer of proliferating neural progenitors. In vertebrates, the entire neurectoderm folds into the embryo to give rise to the neural tube. In Drosophila and many other invertebrates, a subset of neurectodermal cells, called neuroblasts (NBs), delaminates and forms the neural primordium inside the embryo where they divide in an asymmetric, stem cell-like mode. The remainder of the neurectodermal cells that stay behind at the surface loose their neurogenic potential and later give rise to the ventral part of the epidermis. The genetic and molecular analysis of the mechanisms controlling specification and proliferation of NBs in the Drosophila embryo, which played a significant part in pioneering the field of modern developmental neurobiology, represents the topic of this review.
Copyright © 2013 Wiley Periodicals, Inc.
Figures


References
-
- Wheeler WM. Neuroblasts in the arthropod embryo. J Morph. 1891;4:337–343.
-
- Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J Embryol Exp Morphol. 1976;35:107–123. - PubMed
-
- Hartenstein V, Campos-Ortega JA. Early neurogenesis in wildtype Drosophila melanogaster. Roux’s Arch Dev Biol. 1984;193:308–325. - PubMed
-
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. - PubMed
-
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

