Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia
- PMID: 24014546
- PMCID: PMC3809527
- DOI: 10.1105/tpc.113.115741
Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia
Abstract
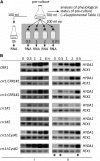
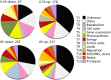
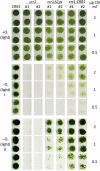
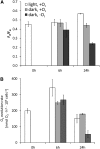
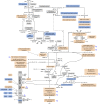
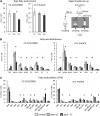
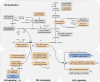
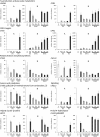
Anaerobiosis is a stress condition for aerobic organisms and requires extensive acclimation responses. We used RNA-Seq for a whole-genome view of the acclimation of Chlamydomonas reinhardtii to anoxic conditions imposed simultaneously with transfer to the dark. Nearly 1.4 × 10(3) genes were affected by hypoxia. Comparing transcript profiles from early (hypoxic) with those from late (anoxic) time points indicated that cells activate oxidative energy generation pathways before employing fermentation. Probable substrates include amino acids and fatty acids (FAs). Lipid profiling of the C. reinhardtii cells revealed that they degraded FAs but also accumulated triacylglycerols (TAGs). In contrast with N-deprived cells, the TAGs in hypoxic cells were enriched in desaturated FAs, suggesting a distinct pathway for TAG accumulation. To distinguish transcriptional responses dependent on copper response regulator1 (CRR1), which is also involved in hypoxic gene regulation, we compared the transcriptomes of crr1 mutants and complemented strains. In crr1 mutants, ~40 genes were aberrantly regulated, reaffirming the importance of CRR1 for the hypoxic response, but indicating also the contribution of additional signaling strategies to account for the remaining differentially regulated transcripts. Based on transcript patterns and previous results, we conclude that nitric oxide-dependent signaling cascades operate in anoxic C. reinhardtii cells.
Figures


References
-
- Adams S., Maple J., Møller S.G. (2008). Functional conservation of the MIN plastid division homologues of Chlamydomonas reinhardtii. Planta 227: 1199–1211 - PubMed
-
- Allen M.D., Kropat J., Merchant S.S. (2008). Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84: 1336–1342 - PubMed
-
- Almeida C.C., Romão C.V., Lindley P.F., Teixeira M., Saraiva L.M. (2006). The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281: 32445–32450 - PubMed
-
- Anbar A.D. (2008). Oceans. Elements and evolution. Science 322: 1481–1483 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

