Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres
- PMID: 24014547
- PMCID: PMC3809539
- DOI: 10.1105/tpc.113.114736
Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres
Abstract
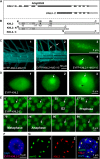
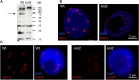
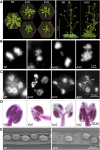
The centromeric histone H3 variant cenH3 is an essential centromeric protein required for assembly, maintenance, and proper function of kinetochores during mitosis and meiosis. We identified a kinetochore null2 (KNL2) homolog in Arabidopsis thaliana and uncovered features of its role in cenH3 loading at centromeres. We show that Arabidopsis KNL2 colocalizes with cenH3 and is associated with centromeres during all stages of the mitotic cell cycle, except from metaphase to mid-anaphase. KNL2 is regulated by the proteasome degradation pathway. The KNL2 promoter is mainly active in meristematic tissues, similar to the cenH3 promoter. A knockout mutant for KNL2 shows a reduced level of cenH3 expression and reduced amount of cenH3 protein at chromocenters of meristematic nuclei, anaphase bridges during mitosis, micronuclei in pollen tetrads, and 30% seed abortion. Moreover, knl2 mutant plants display reduced expression of suppressor of variegation 3-9 homologs2, 4, and 9 and reduced DNA methylation, suggesting an impact of KNL2 on the epigenetic environment for centromere maintenance.
Figures

References
-
- Aasland R., Stewart A.F., Gibson T. (1996). The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21: 87–88 - PubMed
-
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 - PubMed
-
- Allshire R.C. (1997). Centromeres, checkpoints and chromatid cohesion. Curr. Opin. Genet. Dev. 7: 264–273 - PubMed
-
- Berckmans B., De Veylder L. (2009). Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 12: 599–605 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

