Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds
- PMID: 24014578
- PMCID: PMC3793072
- DOI: 10.1104/pp.113.226761
Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds
Abstract
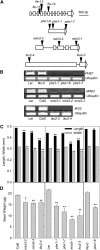
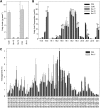
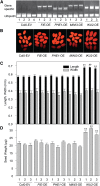
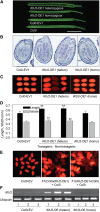
Seed endosperm development in Arabidopsis (Arabidopsis thaliana) is under control of the polycomb group complex, which includes Fertilization Independent Endosperm (FIE). The polycomb group complex regulates downstream factors, e.g. Pheres1 (PHE1), by genomic imprinting. In heterozygous fie mutants, an endosperm develops in ovules carrying a maternal fie allele without fertilization, finally leading to abortion. Another endosperm development pathway depends on MINISEED3 (a WRKY10 transcription factor) and HAIKU2 (a leucine-rich repeat kinase). While the role of seed development genes in the embryo and endosperm establishment has been studied in detail, their impact on metabolism and oil accumulation remained unclear. Analysis of oil, protein, and sucrose accumulation in mutants and overexpression plants of the four seed development genes revealed that (1) seeds carrying a maternal fie allele accumulate low oil with an altered composition of triacylglycerol molecular species; (2) homozygous mutant seeds of phe1, mini3, and iku2, which are smaller, accumulate less oil and slightly less protein, and starch, which accumulates early during seed development, remains elevated in mutant seeds; (3) embryo-specific overexpression of FIE, PHE1, and MINI3 has no influence on seed size and weight, nor on oil, protein, or sucrose content; and (4) overexpression of IKU2 results in seeds with increased size and weight, and oil content of overexpressed IKU2 seeds is increased by 35%. Thus, IKU2 overexpression represents a novel strategy for the genetic manipulation of the oil content in seeds.
Figures





Similar articles
-
MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis.Proc Natl Acad Sci U S A. 2005 Nov 29;102(48):17531-6. doi: 10.1073/pnas.0508418102. Epub 2005 Nov 17. Proc Natl Acad Sci U S A. 2005. PMID: 16293693 Free PMC article.
-
SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development.Plant Cell. 2009 Jan;21(1):106-17. doi: 10.1105/tpc.108.064972. Epub 2009 Jan 13. Plant Cell. 2009. PMID: 19141706 Free PMC article.
-
SHORT HYPOCOTYL UNDER BLUE1 or HAIKU2 mixepression alters canola and Arabidopsis seed development.New Phytol. 2016 Jan;209(2):636-49. doi: 10.1111/nph.13632. Epub 2015 Sep 21. New Phytol. 2016. PMID: 26389843
-
[Genomic imprinting and seed development].Yi Chuan. 2005 Jul;27(4):665-70. Yi Chuan. 2005. PMID: 16120596 Review. Chinese.
-
Epigenetic mechanisms governing seed development in plants.EMBO Rep. 2006 Dec;7(12):1223-7. doi: 10.1038/sj.embor.7400854. EMBO Rep. 2006. PMID: 17139298 Free PMC article. Review.
Cited by
-
Imprinting in plants as a mechanism to generate seed phenotypic diversity.Front Plant Sci. 2015 Jan 27;5:780. doi: 10.3389/fpls.2014.00780. eCollection 2014. Front Plant Sci. 2015. PMID: 25674092 Free PMC article. Review.
-
Comparative transcriptome analysis of cultivated and wild seeds of Salvia hispanica (chia).Sci Rep. 2019 Jul 5;9(1):9761. doi: 10.1038/s41598-019-45895-5. Sci Rep. 2019. PMID: 31278279 Free PMC article.
-
Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis.Plant Physiol. 2014 May;165(1):30-6. doi: 10.1104/pp.114.236430. Epub 2014 Apr 2. Plant Physiol. 2014. PMID: 24696520 Free PMC article.
-
Adaptive evolution of seed oil content in angiosperms: accounting for the global patterns of seed oils.BMC Evol Biol. 2016 Sep 9;16(1):187. doi: 10.1186/s12862-016-0752-7. BMC Evol Biol. 2016. PMID: 27613109 Free PMC article.
-
Molecular Network for Regulation of Seed Size in Plants.Int J Mol Sci. 2023 Jun 26;24(13):10666. doi: 10.3390/ijms241310666. Int J Mol Sci. 2023. PMID: 37445843 Free PMC article. Review.
References
-
- Baroux C, Pien S, Grossniklaus U. (2007) Chromatin modification and remodeling during early seed development. Curr Opin Genet Dev 17: 473–479 - PubMed
-
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160
-
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C. (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 - PubMed
-
- Browse J, McCourt PJ, Somerville CR. (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 - PubMed
-
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

