Efficient nonmeiotic allele introgression in livestock using custom endonucleases
- PMID: 24014591
- PMCID: PMC3799378
- DOI: 10.1073/pnas.1310478110
Efficient nonmeiotic allele introgression in livestock using custom endonucleases
Abstract
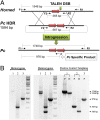
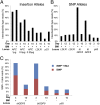
We have expanded the livestock gene editing toolbox to include transcription activator-like (TAL) effector nuclease (TALEN)- and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-stimulated homology-directed repair (HDR) using plasmid, rAAV, and oligonucleotide templates. Toward the genetic dehorning of dairy cattle, we introgressed a bovine POLLED allele into horned bull fibroblasts. Single nucleotide alterations or small indels were introduced into 14 additional genes in pig, goat, and cattle fibroblasts using TALEN mRNA and oligonucleotide transfection with efficiencies of 10-50% in populations. Several of the chosen edits mimic naturally occurring performance-enhancing or disease- resistance alleles, including alteration of single base pairs. Up to 70% of the fibroblast colonies propagated without selection harbored the intended edits, of which more than one-half were homozygous. Edited fibroblasts were used to generate pigs with knockout alleles in the DAZL and APC genes to model infertility and colon cancer. Our methods enable unprecedented meiosis-free intraspecific and interspecific introgression of select alleles in livestock for agricultural and biomedical applications.
Conflict of interest statement
Conflict of interest statement: D.F.C., C.A.L., D.A.W., P.B.H., and S.C.F. either have equity in and/or work for Recombinetics, Inc., a biotechnology company that receives funding from the National Institutes of Health to develop gene editing in commercial livestock.
Figures



Comment in
-
Natural genotypes via genetic engineering.Proc Natl Acad Sci U S A. 2013 Oct 8;110(41):16295-6. doi: 10.1073/pnas.1315623110. Epub 2013 Sep 27. Proc Natl Acad Sci U S A. 2013. PMID: 24077259 Free PMC article. No abstract available.
References
-
- Hammer RE, et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315(6021):680–683. - PubMed
-
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23(8):967–973. - PubMed
-
- Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

