Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells
- PMID: 24014651
- PMCID: PMC3805851
- DOI: 10.1210/me.2013-1077
Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells
Abstract
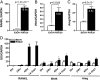
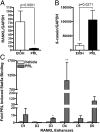
Progesterone (P4) stimulates proliferation of the mammary epithelium by a mechanism that involves paracrine signaling mediated from progesterone receptor (PR)-positive to neighboring PR-negative cells. Here we used a primary mouse mammary epithelial cell (MEC) culture system to define the molecular mechanism by which P4 regulates the expression of target gene effectors of proliferation including the paracrine factor receptor and activator of nuclear factor κB ligand (RANKL). MECs from adult virgin mice grown and embedded in three-dimensional basement-membrane medium resemble mammary ducts in vivo structurally and with respect to other properties including a heterogeneous pattern of PR expression, P4 induction of RANKL and other target genes in a PR-dependent manner, and a proliferative response to progestin. RANKL was demonstrated to have multiple functional P4-responsive enhancers that bind PR in a hormone-dependent manner as detected by chromatin immunoprecipitation assay. P4 also stimulated recruitment of signal transducer and activator of transcription (Stat)5a to RANKL enhancers through an apparent tethering with PR. Analysis of primary MECs from Stat5a knockout mice revealed that P4 induction of RANKL and a broad range of other PR target genes required Stat5a, as did P4-stimulated cell proliferation. In the absence of Stat5a, PR binding was lost at selective RANKL enhancers but was retained with others, suggesting that Stat5a acts to facilitate PR DNA binding at selective sites and to function as a coactivator with DNA-bound PR at others. These results show that RANKL is a direct PR target gene and that Stat5a has a novel role as a cofactor in PR-mediated transcriptional signaling in the mammary gland.
Figures







References
-
- Medina D. The mammary gland: a unique organ for the study of development and tumorigenesis. J Mammary Gland Biol Neoplasia. 1996;1:5–19 - PubMed
-
- Fendrick JL, Raafat AM, Haslam SZ. Mammary gland growth and development from the postnatal period to postmenopause: ovarian steroid receptor ontogeny and regulation in the mouse. J Mammary Gland Biol Neoplasia. 1998;3:7–22 - PubMed
-
- Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289:1751–1754 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

