Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation
- PMID: 24015111
- PMCID: PMC3754893
- DOI: 10.1371/journal.pbio.1001643
Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation
Abstract
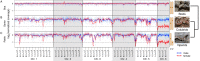
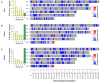
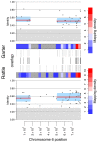
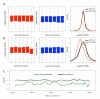
Snakes exhibit genetic sex determination, with female heterogametic sex chromosomes (ZZ males, ZW females). Extensive cytogenetic work has suggested that the level of sex chromosome heteromorphism varies among species, with Boidae having entirely homomorphic sex chromosomes, Viperidae having completely heteromorphic sex chromosomes, and Colubridae showing partial differentiation. Here, we take a genomic approach to compare sex chromosome differentiation in these three snake families. We identify homomorphic sex chromosomes in boas (Boidae), but completely heteromorphic sex chromosomes in both garter snakes (Colubridae) and pygmy rattlesnake (Viperidae). Detection of W-linked gametologs enables us to establish the presence of evolutionary strata on garter and pygmy rattlesnake sex chromosomes where recombination was abolished at different time points. Sequence analysis shows that all strata are shared between pygmy rattlesnake and garter snake, i.e., recombination was abolished between the sex chromosomes before the two lineages diverged. The sex-biased transmission of the Z and its hemizygosity in females can impact patterns of molecular evolution, and we show that rates of evolution for Z-linked genes are increased relative to their pseudoautosomal homologs, both at synonymous and amino acid sites (even after controlling for mutational biases). This demonstrates that mutation rates are male-biased in snakes (male-driven evolution), but also supports faster-Z evolution due to differential selective effects on the Z. Finally, we perform a transcriptome analysis in boa and pygmy rattlesnake to establish baseline levels of sex-biased expression in homomorphic sex chromosomes, and show that heteromorphic ZW chromosomes in rattlesnakes lack chromosome-wide dosage compensation. Our study provides the first full scale overview of the evolution of snake sex chromosomes at the genomic level, thus greatly expanding our knowledge of reptilian and vertebrate sex chromosomes evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Bull JJ (1983) Evolution of sex determining mechanisms. Menlo Park (California): Benjamin Cummings.
-
- Charlesworth B (1996) The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6: 149–162. - PubMed
-
- Rice WR (1987) The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 41: 911–914. - PubMed

