Retinal connectomics: towards complete, accurate networks
- PMID: 24016532
- PMCID: PMC4045117
- DOI: 10.1016/j.preteyeres.2013.08.002
Retinal connectomics: towards complete, accurate networks
Abstract
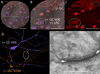
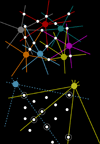
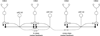
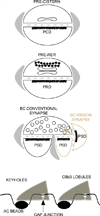
Connectomics is a strategy for mapping complex neural networks based on high-speed automated electron optical imaging, computational assembly of neural data volumes, web-based navigational tools to explore 10(12)-10(15) byte (terabyte to petabyte) image volumes, and annotation and markup tools to convert images into rich networks with cellular metadata. These collections of network data and associated metadata, analyzed using tools from graph theory and classification theory, can be merged with classical systems theory, giving a more completely parameterized view of how biologic information processing systems are implemented in retina and brain. Networks have two separable features: topology and connection attributes. The first findings from connectomics strongly validate the idea that the topologies of complete retinal networks are far more complex than the simple schematics that emerged from classical anatomy. In particular, connectomics has permitted an aggressive refactoring of the retinal inner plexiform layer, demonstrating that network function cannot be simply inferred from stratification; exposing the complex geometric rules for inserting different cells into a shared network; revealing unexpected bidirectional signaling pathways between mammalian rod and cone systems; documenting selective feedforward systems, novel candidate signaling architectures, new coupling motifs, and the highly complex architecture of the mammalian AII amacrine cell. This is but the beginning, as the underlying principles of connectomics are readily transferrable to non-neural cell complexes and provide new contexts for assessing intercellular communication.
Keywords: Connectome; Gap junctions; Networks; Neurons; Retina; Synapses.
Copyright © 2013 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures














References
-
- Amari SI, Beltrame F, Bjaalie JG, Dalkara T, Schutter ED, Egan GF, Goddard NH, Gonzalez C, Grillner S, Herz A, Hoffmann KP, Jaaskelainen I, Koslow SH, Lee SY, Matthiessen L, Miller PL, Silva FMD, Novak M, Ravindranath V, Ritz R, Ruotsalainen U, Sebestra V, Subramaniam S, Tang Y, Toga AW, Usui S, Pelt JV, Verschure P, Willshaw D, Wrobel A. Neuroinformatics: The integration of shared databases and tools towards integrative neuroscience. Journal of Integrative Neuroscience. 2002;1:117–128. - PubMed
-
- Aster R, Borchers B, Thurber C. Parameter Estimation and Inverse. Problems. NY: Academic Press; 2005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

