Flavodoxin cofactor binding induces structural changes that are required for protein-protein interactions with NADP(+) oxidoreductase and pyruvate formate-lyase activating enzyme
- PMID: 24016774
- PMCID: PMC4012331
- DOI: 10.1016/j.bbapap.2013.08.014
Flavodoxin cofactor binding induces structural changes that are required for protein-protein interactions with NADP(+) oxidoreductase and pyruvate formate-lyase activating enzyme
Abstract
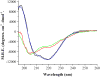
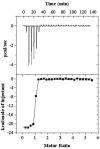
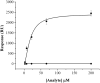
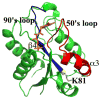
Flavodoxin (Fld) conformational changes, thermal stability, and cofactor binding were studied using circular dichroism (CD), isothermal titration calorimetry (ITC), and limited proteolysis. Thermodynamics of apo and holo-Fld folding were examined to discern the features of this important electron transfer protein and to provide data on apo-Fld. With the exception of fluorescence and UV-vis binding experiments with its cofactor flavin mononucleotide (FMN), apo-Fld is almost completely uncharacterized in Escherichia coli. Fld is more structured when the FMN cofactor is bound; the association is tight and driven by enthalpy of binding. Surface plasmon resonance binding experiments were carried out under anaerobic conditions for both apo- and holo-Fld and demonstrate the importance of structure and conformation for the interaction with binding partners. Holo-Fld is capable of associating with NADP(+)-dependent flavodoxin oxidoreductase (FNR) and pyruvate formate-lyase activating enzyme (PFL-AE) whereas there is no detectable interaction between apo-Fld and either protein. Limited proteolysis experiments were analyzed by LC-MS to identify the regions in Fld that are involved in conformation changes upon cofactor binding. Docking software was used to model the Fld/PFL-AE complex to understand the interactions between these two proteins and gain insight into electron transfer reactions from Fld to PFL-AE.
Keywords: Circular dichroism (CD); Flavodoxin; NADP(+) oxidoreductase; Protein–protein interactions; Pyruvate formate-lyase activating enzyme; Surface plasmon resonance (SPR).
© 2013.
Figures



Similar articles
-
Flavodoxin-mediated electron transfer from photosystem I to ferredoxin-NADP+ reductase in Anabaena: role of flavodoxin hydrophobic residues in protein-protein interactions.Biochemistry. 2008 Jan 29;47(4):1207-17. doi: 10.1021/bi7017392. Epub 2008 Jan 5. Biochemistry. 2008. PMID: 18177021
-
Effects of chemical modification of Anabaena flavodoxin and ferredoxin-NADP+ reductase on the kinetics of interprotein electron transfer reactions.Eur J Biochem. 1992 Dec 1;210(2):577-83. doi: 10.1111/j.1432-1033.1992.tb17457.x. Eur J Biochem. 1992. PMID: 1459139
-
A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.Biochem Biophys Res Commun. 2023 Jan 8;639:134-141. doi: 10.1016/j.bbrc.2022.11.089. Epub 2022 Nov 29. Biochem Biophys Res Commun. 2023. PMID: 36493556
-
Evolution of the acceptor side of photosystem I: ferredoxin, flavodoxin, and ferredoxin-NADP+ oxidoreductase.Photosynth Res. 2017 Dec;134(3):235-250. doi: 10.1007/s11120-017-0338-2. Epub 2017 Feb 1. Photosynth Res. 2017. PMID: 28150152 Review.
-
P450BM-3; a tale of two domains--or is it three?Steroids. 1997 Jan;62(1):117-23. doi: 10.1016/s0039-128x(96)00169-9. Steroids. 1997. PMID: 9029725 Review.
Cited by
-
C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products.Nat Prod Rep. 2018 Jul 18;35(7):660-694. doi: 10.1039/c8np00006a. Nat Prod Rep. 2018. PMID: 29633774 Free PMC article. Review.
-
Glycyl radical activating enzymes: structure, mechanism, and substrate interactions.Arch Biochem Biophys. 2014 Mar 15;546:64-71. doi: 10.1016/j.abb.2014.01.020. Epub 2014 Jan 31. Arch Biochem Biophys. 2014. PMID: 24486374 Free PMC article. Review.
-
Glycyl Radical Enzyme-Associated Microcompartments: Redox-Replete Bacterial Organelles.mBio. 2019 Jan 8;10(1):e02327-18. doi: 10.1128/mBio.02327-18. mBio. 2019. PMID: 30622187 Free PMC article. Review.
-
A cellular selection identifies elongated flavodoxins that support electron transfer to sulfite reductase.Protein Sci. 2023 Oct;32(10):e4746. doi: 10.1002/pro.4746. Protein Sci. 2023. PMID: 37551563 Free PMC article.
-
Radical S-adenosylmethionine enzymes.Chem Rev. 2014 Apr 23;114(8):4229-317. doi: 10.1021/cr4004709. Epub 2014 Jan 29. Chem Rev. 2014. PMID: 24476342 Free PMC article. Review. No abstract available.
References
-
- Fukuyama K, Matsubara H, Rodgers LJ. Crystal structure of oxidized flavodoxin from red algae Chlondrus crispus refined at 1.8 Å resolution. Description of the flavodoxin mononucleotide binding site. J Mol Biol. 1992;225:775–789. - PubMed
-
- Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J Biol Chem. 2001;276:35558–35563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

