Isolevuglandin-modified phosphatidylethanolamine is metabolized by NAPE-hydrolyzing phospholipase D
- PMID: 24018423
- PMCID: PMC3793619
- DOI: 10.1194/jlr.M042556
Isolevuglandin-modified phosphatidylethanolamine is metabolized by NAPE-hydrolyzing phospholipase D
Abstract
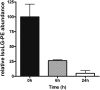
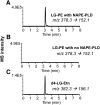
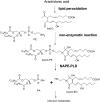
Lipid aldehydes including isolevuglandins (IsoLGs) and 4-hydroxynonenal modify phosphatidylethanolamine (PE) to form proinflammatory and cytotoxic adducts. Therefore, cells may have evolved mechanisms to degrade and prevent accumulation of these potentially harmful compounds. To test if cells could degrade isolevuglandin-modified phosphatidylethanolamine (IsoLG-PE), we generated IsoLG-PE in human embryonic kidney 293 (HEK293) cells and human umbilical cord endothelial cells and measured its stability over time. We found that IsoLG-PE levels decreased more than 75% after 6 h, suggesting that IsoLG-PE was indeed degraded. Because N-acyl phosphatidylethanolamine-hydrolyzing phospholipase D (NAPE-PLD) has been described as a key enzyme in the hydrolysis of N-acyl phosphatidylethanoamine (NAPE) and both NAPE and IsoLG-PE have large aliphatic headgroups, we considered the possibility that this enzyme might also hydrolyze IsoLG-PE. We found that knockdown of NAPE-PLD expression using small interfering RNA (siRNA) significantly increased the persistence of IsoLG-PE in HEK293 cells. IsoLG-PE competed with NAPE for hydrolysis by recombinant mouse NAPE-PLD, with the catalytic efficiency (V(max)/K(m)) for hydrolysis of IsoLG-PE being 30% of that for hydrolysis of NAPE. LC-MS/MS analysis confirmed that recombinant NAPE-PLD hydrolyzed IsoLG-PE to IsoLG-ethanolamine. These results demonstrate that NAPE-PLD contributes to the degradation of IsoLG-PE and suggest that a major physiological role of NAPE-PLD may be to degrade aldehyde-modified PE, thereby preventing the accumulation of these harmful compounds.
Keywords: N-acyl ethanolamine; N-acyl phosphatidylethanolamine; cytotoxicity; inflammation; isoketal; oxidative stress.
Figures





Similar articles
-
N-aldehyde-modified phosphatidylethanolamines generated by lipid peroxidation are robust substrates of N-acyl phosphatidylethanolamine phospholipase D.J Lipid Res. 2025 Jul;66(7):100831. doi: 10.1016/j.jlr.2025.100831. Epub 2025 May 21. J Lipid Res. 2025. PMID: 40409473 Free PMC article.
-
N-Aldehyde-Modified Phosphatidylethanolamines generated by lipid peroxidation are robust substrates of N-Acyl Phosphatidylethanolamine Phospholipase D.bioRxiv [Preprint]. 2024 Nov 1:2024.10.30.621135. doi: 10.1101/2024.10.30.621135. bioRxiv. 2024. Update in: J Lipid Res. 2025 Jul;66(7):100831. doi: 10.1016/j.jlr.2025.100831. PMID: 39554116 Free PMC article. Updated. Preprint.
-
Peripheral tissue levels and molecular species compositions of N-acyl-phosphatidylethanolamine and its metabolites in mice lacking N-acyl-phosphatidylethanolamine-specific phospholipase D.J Biochem. 2017 Dec 1;162(6):449-458. doi: 10.1093/jb/mvx054. J Biochem. 2017. PMID: 28992041
-
Endocannabinoid-related enzymes as drug targets with special reference to N-acylphosphatidylethanolamine-hydrolyzing phospholipase D.Curr Med Chem. 2005;12(12):1413-22. doi: 10.2174/0929867054020918. Curr Med Chem. 2005. PMID: 15974992 Review.
-
Enzymatic formation of anandamide.Vitam Horm. 2009;81:1-24. doi: 10.1016/S0083-6729(09)81001-7. Vitam Horm. 2009. PMID: 19647106 Review.
Cited by
-
Functional Genomic Analysis of the Impact of Camelina (Camelina sativa) Meal on Atlantic Salmon (Salmo salar) Distal Intestine Gene Expression and Physiology.Mar Biotechnol (NY). 2016 Jun;18(3):418-35. doi: 10.1007/s10126-016-9704-x. Epub 2016 Jun 2. Mar Biotechnol (NY). 2016. PMID: 27255337 Free PMC article.
-
Symmetrically substituted dichlorophenes inhibit N-acyl-phosphatidylethanolamine phospholipase D.J Biol Chem. 2020 May 22;295(21):7289-7300. doi: 10.1074/jbc.RA120.013362. Epub 2020 Apr 13. J Biol Chem. 2020. PMID: 32284327 Free PMC article.
-
Isolevuglandin adducts in disease.Antioxid Redox Signal. 2015 Jun 20;22(18):1703-18. doi: 10.1089/ars.2014.6154. Epub 2015 Feb 18. Antioxid Redox Signal. 2015. PMID: 25557218 Free PMC article. Review.
-
Discovery of desketoraloxifene analogues as inhibitors of mammalian, Pseudomonas aeruginosa, and NAPE phospholipase D enzymes.ACS Chem Biol. 2015 Feb 20;10(2):421-32. doi: 10.1021/cb500828m. Epub 2014 Nov 19. ACS Chem Biol. 2015. PMID: 25384256 Free PMC article.
-
Isolevuglandin-type lipid aldehydes induce the inflammatory response of macrophages by modifying phosphatidylethanolamines and activating the receptor for advanced glycation endproducts.Antioxid Redox Signal. 2015 Jun 20;22(18):1633-45. doi: 10.1089/ars.2014.6078. Epub 2015 Mar 18. Antioxid Redox Signal. 2015. PMID: 25751734 Free PMC article.
References
-
- Salomon R. G., Miller D. B., Zagorski M. G., Coughlin D. J. 1984. Solvent-induced fragmentation of prostaglandin endoperoxides. New aldehyde products from PGH2 and a novel intramolecular 1,2-hydride shift during endoperoxide fragmentation in aqueous solution. J. Am. Chem. Soc. 106: 6049–6060
-
- Salomon R. G., Miller D. B. 1985. Levuglandins: isolation, characterization, and total synthesis of new secoprostanoid products from prostaglandin endoperoxides. Adv. Prostaglandin Thromboxane Leukot. Res. 15: 323–326 - PubMed
-
- Fukuda K., Davies S. S., Nakajima T., Ong B. H., Kupershmidt S., Fessel J., Amarnath V., Anderson M. E., Boyden P. A., Viswanathan P. C., et al. 2005. Oxidative mediated lipid peroxidation recapitulates proarrhythmic effects on cardiac sodium channels. Circ. Res. 97: 1262–1269 - PubMed
-
- Hoppe G., Subbanagounder G., O'Neil J., Salomon R. G., Hoff H. F. 1997. Macrophage recognition of LDL modified by levuglandin E2, an oxidation product of arachidonic acid. Biochim. Biophys. Acta. 1344: 1–5 - PubMed
-
- Bernoud-Hubac N., Alam D. A., Lefils J., Davies S. S., Amarnath V., Guichardant M., Roberts L. J., 2nd, Lagarde M. 2009. Low concentrations of reactive gamma-ketoaldehydes prime thromboxane-dependent human platelet aggregation via p38-MAPK activation. Biochim. Biophys. Acta. 1791: 307–313 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

