Major histocompatibility complex class I core promoter elements are not essential for transcription in vivo
- PMID: 24019072
- PMCID: PMC3838175
- DOI: 10.1128/MCB.00553-13
Major histocompatibility complex class I core promoter elements are not essential for transcription in vivo
Abstract
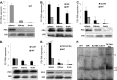
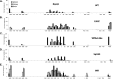
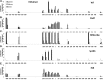
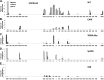
The role of core promoter elements in regulating transcription initiation is largely unknown for genes subject to complex regulation. Major histocompatibility complex class I genes are ubiquitously expressed and governed by tissue-specific and hormonal signals. Transcription initiates at multiple sites within the core promoter, which contains elements homologous to the canonical elements CCAAT, TATAA, Sp1 binding site (Sp1BS), and Initiator (Inr). To determine their functions, expression of class I transgenes with individually mutated elements was assessed. Surprisingly, all mutant promoters supported transcription. However, each mutated core promoter element had a distinct effect on expression: CAAT box mutations modulated constitutive expression in nonlymphoid tissues, whereas TATAA-like element mutations dysregulated transcription in lymphoid tissues. Inr mutations aberrantly elevated expression. Sp1BS element mutations resulted in variegated transgene expression. RNA polymerase II binding and histone H3K4me3 patterns correlated with transgene expression; H3K9me3 marks partially correlated. Whereas the wild-type, TATAA-like, and CAAT mutant promoters were activated by gamma interferon, the Sp1 and Inr mutants were repressed, implicating these elements in regulation of hormonal responses. These results lead to the surprising conclusion that no single element is required for promoter activity. Rather, each plays a distinct role in promoter activity, chromatin structure, tissue-specific expression, and extracellular signaling.
Figures


References
-
- Berk AJ. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330–335 - PubMed
-
- Roeder RG. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327–335 - PubMed
-
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93–105 - PubMed
-
- Gill G. 2001. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 37:33–43 - PubMed
-
- Kadonaga JT. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116:247–257 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
