Transcriptional control by two interacting regulatory proteins: identification of the PtxS binding site at PtxR
- PMID: 24019239
- PMCID: PMC3905896
- DOI: 10.1093/nar/gkt773
Transcriptional control by two interacting regulatory proteins: identification of the PtxS binding site at PtxR
Abstract
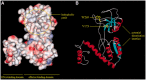
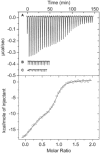
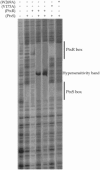
The PtxS and PtxR regulators control the expression of the glucose dehydrogenase genes from the Pgad promoter in Pseudomonas aeruginosa. These regulators bind to their cognate operators, that are separated by ∼50 nt, within the promoter region and interact with each other creating a DNA-loop that prevents RNA polymerase promoter access. Binding of the 2-ketogluconate effector to PtxS caused PtxS/PtxR complex dissociation and led to the dissolution of the repression loop facilitating the entry of the RNA polymerase and enabling the transcription of the gad gene. We have identified a hydrophobic surface patch on the PtxR putative surface that was hypothesized to correspond to the binding site for PtxS. Two surface-exposed residues in this patch, V173 and W269, were replaced by alanine. Isothermal titration calorimetry assays showed that PtxS does not interact with the mutant variants of PtxR. Electrophoretic mobility shift assay and DNAase I footprinting assays proved that both regulators bind to their target operators and that failure to interact with each other prevented the formation of the DNA-loop. In vitro transcription showed that PtxS per se is sufficient to inhibit transcription from the Pgad promoter, but that affinity of PtxS for its effector is modulated by PtxR.
Figures


References
-
- Cao H, Baldini RL, Rahme LG. Common mechanisms for pathogens of plants and animals. Annu. Rev. Phytopathol. 2001;39:259–284. - PubMed
-
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. - PubMed
-
- Hamood AN, Colmer HJ, Carty NL. Regulation of Pseudomonas aeruginosa exotoxin A synthesis. In: Ramos JL, editor. Pseudomonas: Virulence and Gene Regulation. Vol. 2. Amsterdam: Plenum Press; 2004. pp. 389–423.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

