Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis
- PMID: 24019478
- PMCID: PMC3799349
- DOI: 10.1073/pnas.1315532110
Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis
Abstract
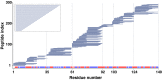
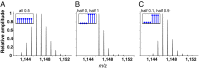
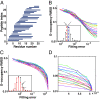
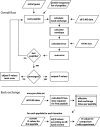
Hydrogen exchange technology provides a uniquely powerful instrument for measuring protein structural and biophysical properties, quantitatively and in a nonperturbing way, and determining how these properties are implemented to produce protein function. A developing hydrogen exchange-mass spectrometry method (HX MS) is able to analyze large biologically important protein systems while requiring only minuscule amounts of experimental material. The major remaining deficiency of the HX MS method is the inability to deconvolve HX results to individual amino acid residue resolution. To pursue this goal we used an iterative optimization program (HDsite) that integrates recent progress in multiple peptide acquisition together with previously unexamined isotopic envelope-shape information and a site-resolved back-exchange correction. To test this approach, residue-resolved HX rates computed from HX MS data were compared with extensive HX NMR measurements, and analogous comparisons were made in simulation trials. These tests found excellent agreement and revealed the important computational determinants.
Keywords: HDX-MS; isotope pattern; protein biophysics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Hydrogen Exchange Mass Spectrometry.Methods Enzymol. 2016;566:335-56. doi: 10.1016/bs.mie.2015.06.035. Epub 2015 Jul 27. Methods Enzymol. 2016. PMID: 26791986 Free PMC article.
-
Native State Hydrogen Exchange-Mass Spectrometry Methods to Probe Protein Folding and Unfolding.Methods Mol Biol. 2022;2376:143-159. doi: 10.1007/978-1-0716-1716-8_8. Methods Mol Biol. 2022. PMID: 34845608
-
High-Resolution Hydrogen-Deuterium Protection Factors from Sparse Mass Spectrometry Data Validated by Nuclear Magnetic Resonance Measurements.J Am Soc Mass Spectrom. 2022 May 4;33(5):813-822. doi: 10.1021/jasms.2c00005. Epub 2022 Apr 6. J Am Soc Mass Spectrom. 2022. PMID: 35385652 Free PMC article.
-
Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
-
Hydrogen exchange and mass spectrometry: A historical perspective.J Am Soc Mass Spectrom. 2006 Nov;17(11):1481-1489. doi: 10.1016/j.jasms.2006.06.006. Epub 2006 Jul 28. J Am Soc Mass Spectrom. 2006. PMID: 16876429 Free PMC article. Review.
Cited by
-
Hydrogen/Deuterium Exchange Mass Spectrometry of Heme-Based Oxygen Sensor Proteins.Methods Mol Biol. 2023;2648:99-122. doi: 10.1007/978-1-0716-3080-8_8. Methods Mol Biol. 2023. PMID: 37039988
-
Mapping residual structure in intrinsically disordered proteins at residue resolution using millisecond hydrogen/deuterium exchange and residue averaging.J Am Soc Mass Spectrom. 2015 Apr;26(4):547-54. doi: 10.1007/s13361-014-1033-6. Epub 2014 Dec 7. J Am Soc Mass Spectrom. 2015. PMID: 25481641
-
Cytochrome c folds through foldon-dependent native-like intermediates in an ordered pathway.Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):3809-14. doi: 10.1073/pnas.1522674113. Epub 2016 Mar 10. Proc Natl Acad Sci U S A. 2016. PMID: 26966231 Free PMC article.
-
Automated Removal of Phospholipids from Membrane Proteins for H/D Exchange Mass Spectrometry Workflows.Anal Chem. 2018 Jun 5;90(11):6409-6412. doi: 10.1021/acs.analchem.8b00429. Epub 2018 May 9. Anal Chem. 2018. PMID: 29723469 Free PMC article.
-
An Allosteric Anti-tryptase Antibody for the Treatment of Mast Cell-Mediated Severe Asthma.Cell. 2019 Oct 3;179(2):417-431.e19. doi: 10.1016/j.cell.2019.09.009. Cell. 2019. PMID: 31585081 Free PMC article.
References
-
- Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17(1):87–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

