Bacteria- and IMD pathway-independent immune defenses against Plasmodium falciparum in Anopheles gambiae
- PMID: 24019865
- PMCID: PMC3760850
- DOI: 10.1371/journal.pone.0072130
Bacteria- and IMD pathway-independent immune defenses against Plasmodium falciparum in Anopheles gambiae
Abstract
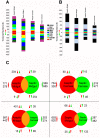
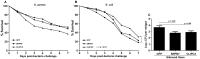
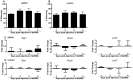
The mosquito Anopheles gambiae uses its innate immune system to control bacterial and Plasmodium infection of its midgut tissue. The activation of potent IMD pathway-mediated anti-Plasmodium falciparum defenses is dependent on the presence of the midgut microbiota, which activate this defense system upon parasite infection through a peptidoglycan recognition protein, PGRPLC. We employed transcriptomic and reverse genetic analyses to compare the P. falciparum infection-responsive transcriptomes of septic and aseptic mosquitoes and to determine whether bacteria-independent anti-Plasmodium defenses exist. Antibiotic treated aseptic mosquitoes mounted molecular immune responses representing a variety of immune functions upon P. falciparum infection. Among other immune factors, our analysis uncovered a serine protease inhibitor (SRPN7) and Clip-domain serine protease (CLIPC2) that were transcriptionally induced in the midgut upon P. falciparum infection, independent of bacteria. We also showed that SRPN7 negatively and CLIPC2 positively regulate the anti-Plasmodium defense, independently of the midgut-associated bacteria. Co-silencing assays suggested that these two genes may function together in a signaling cascade. Neither gene was regulated, nor modulated, by infection with the rodent malaria parasite Plasmodium berghei, suggesting that SRPN7 and CLIPC2 are components of a defense system with preferential activity towards P. falciparum. Further analysis using RNA interference determined that these genes do not regulate the anti-Plasmodium defense mediated by the IMD pathway, and both factors act as agonists of the endogenous midgut microbiota, further demonstrating the lack of functional relatedness between these genes and the bacteria-dependent activation of the IMD pathway. This is the first study confirming the existence of a bacteria-independent, anti-P. falciparum defense. Further exploration of this anti-Plasmodium defense will help clarify determinants of immune specificity in the mosquito, and expose potential gene and/or protein targets for malaria intervention strategies based on targeting the parasite in the mosquito vector.
Conflict of interest statement
Figures



References
-
- Gulland A (2012) Death toll from malaria is double the WHO estimate, study finds. BMJ 344: e895. - PubMed
-
- Meister S, Koutsos AC, Christophides GK (2004) The Plasmodium parasite–a 'new' challenge for insect innate immunity. Int J Parasitol 34: 1473–1482. - PubMed
-
- Michel K, Kafatos FC (2005) Mosquito immunity against Plasmodium. Insect Biochem Mol Biol 35: 677–689. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

