Geographically structured populations of Cryptococcus neoformans Variety grubii in Asia correlate with HIV status and show a clonal population structure
- PMID: 24019866
- PMCID: PMC3760895
- DOI: 10.1371/journal.pone.0072222
Geographically structured populations of Cryptococcus neoformans Variety grubii in Asia correlate with HIV status and show a clonal population structure
Abstract
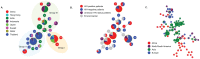
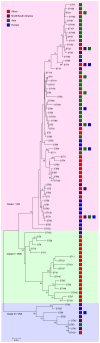
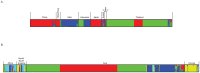
Cryptococcosis is an important fungal disease in Asia with an estimated 140,000 new infections annually the majority of which occurs in patients suffering from HIV/AIDS. Cryptococcus neoformans variety grubii (serotype A) is the major causative agent of this disease. In the present study, multilocus sequence typing (MLST) using the ISHAM MLST consensus scheme for the C. neoformans/C. gattii species complex was used to analyse nucleotide polymorphisms among 476 isolates of this pathogen obtained from 8 Asian countries. Population genetic analysis showed that the Asian C. neoformans var. grubii population shows limited genetic diversity and demonstrates a largely clonal mode of reproduction when compared with the global MLST dataset. HIV-status, sequence types and geography were found to be confounded. However, a correlation between sequence types and isolates from HIV-negative patients was observed among the Asian isolates. Observations of high gene flow between the Middle Eastern and the Southeastern Asian populations suggest that immigrant workers in the Middle East were originally infected in Southeastern Asia.
Conflict of interest statement
Figures

References
-
- Bovers M, Hagen F, Boekhout T (2008) Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev Iberoam Micol 25: S4–12. - PubMed
-
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530. - PubMed
-
- Banerjee U, Datta K, Casadevall A (2004) Serotype distribution of Cryptococcus neoformans in patients in a tertiary care center in India. Med Mycol 42: 181–186. - PubMed
-
- Ganiem AR, Parwati I, Wisaksana R, van der Zanden A, van de Beek D, et al. (2009) The effect of HIV infection on adult meningitis in Indonesia: a prospective cohort study. AIDS 23: 2309–2316. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

